¶ Solid-Phase Synthesis
When synthesizing biopolymers, a lot of steps will be involved. It is therefore crucial for the synthetic process to fufill the following requirements
- High yield reaction steps
- High excess of reagents to help increase the yield
- Cheap building block, so it is not costly to use a high excess of reagents
- Fast reactions
These conditions can be satisfied with Solid-Phase Synthesis
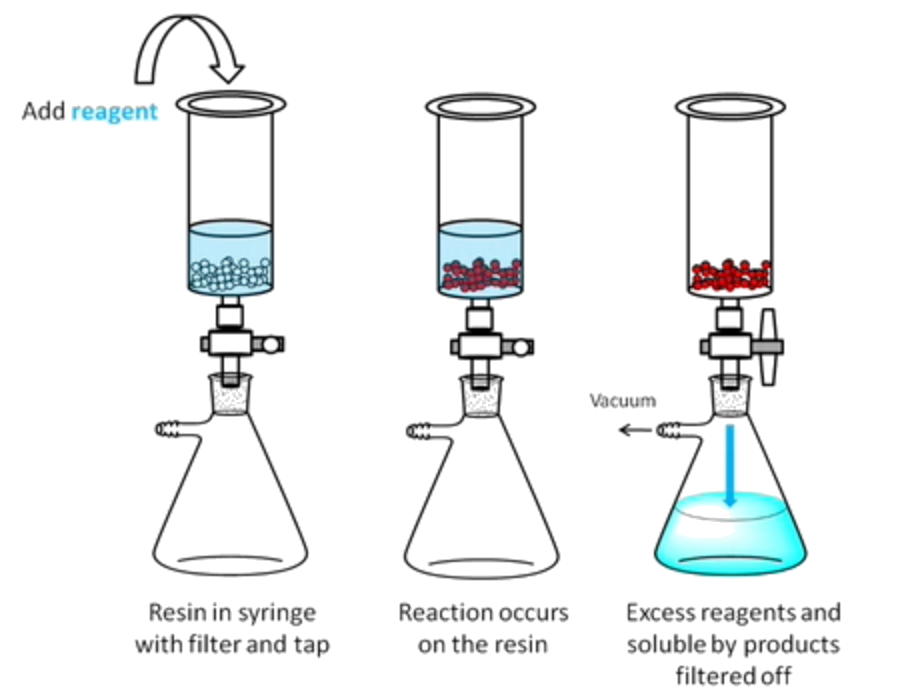
Principle of Solid-Phase Synthesis
We immobilize our reactant on a solid-support, called a resin
- In biopolymers, this is where we grow our polymer chain
We put our resin in a vessel such that when a vaccum is applied, reagents that are not attached to the resin can be removed
- The resin is washed with solvent in between each step to ensure the excess reagents are removed
- No work-up or purification is needed in between each steps, which results in a higher yield
- Unlike solution synthesis, we do multiple steps in a row without characterization
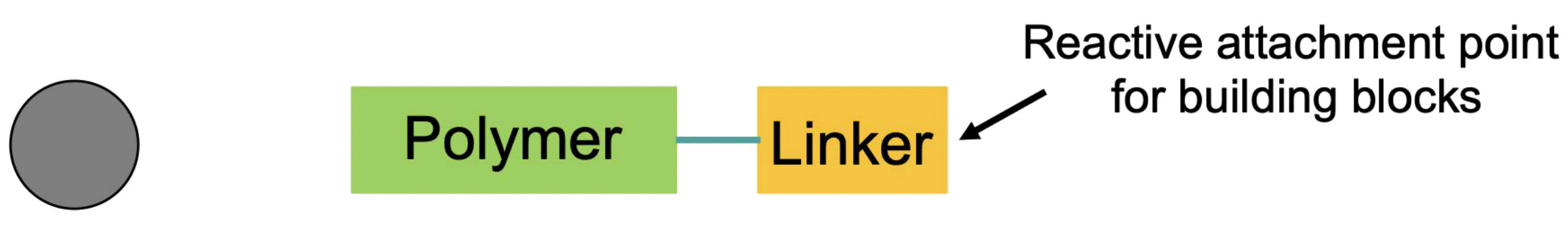
Resins
Resin beads are usually made of inert polymers like polystyrene or polyamide
- The beads must be inert to all the coupling reagents that will be used in the reaction
- The bead must swell in solvent so that it can allow reagents to get through the holes to come into contact with the reactants that have been attached to the resin
- The resin polymer must be functionalized with a reactive handle ( aka linker ) to allow us to immobilize our first reagent to the resin
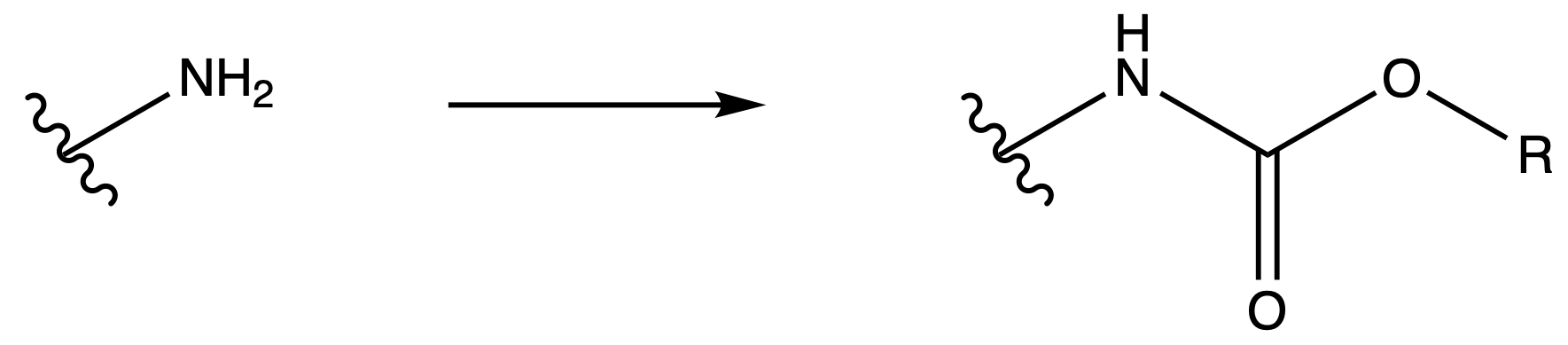
¶ Protecting Groups
Protecting groups temporarily mask specific functional groups in a molecule so other functional groups can be selectively transformed
- Protecting groups are commonly used when a specific functional group is vulnerable to the reagents or conditions necessary for a desired transformation
- The protecting group is then removed later in the synthesis to reveal the original functional group.
¶ Carboxylic Acids
We want to protect the carboxyl group so as to
- Avoid proton donation
- Nucleophilic attack
- Enolisation
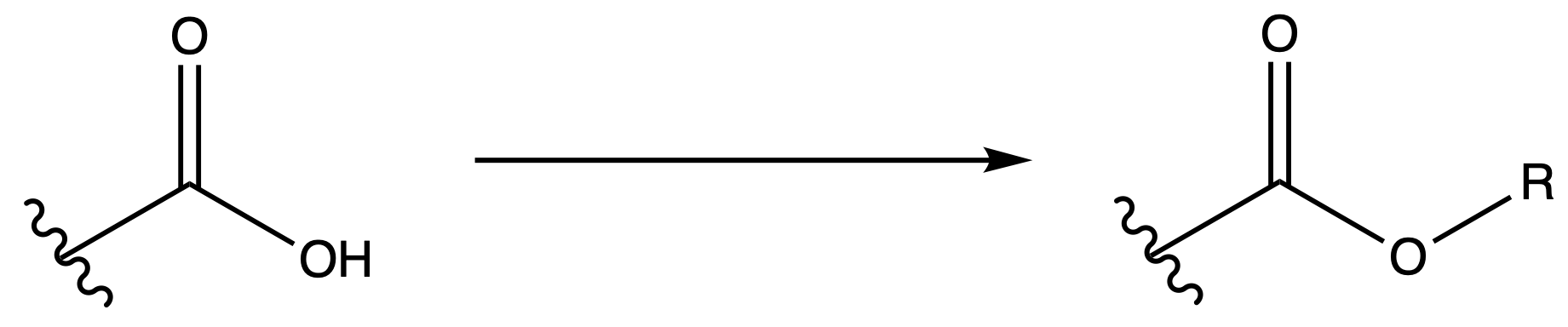
We can achieve this by converting the carboxyl group into a bulky ester group such that the steric hinderance will kinetically prevent reactions
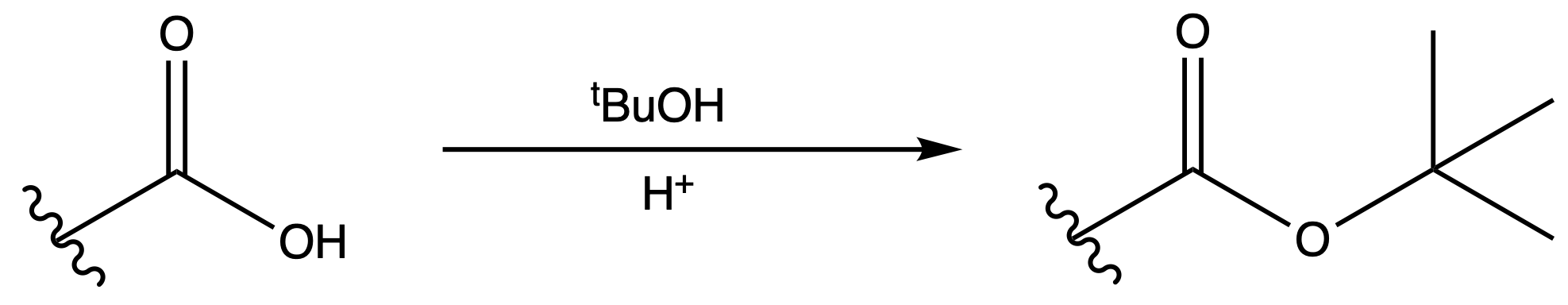
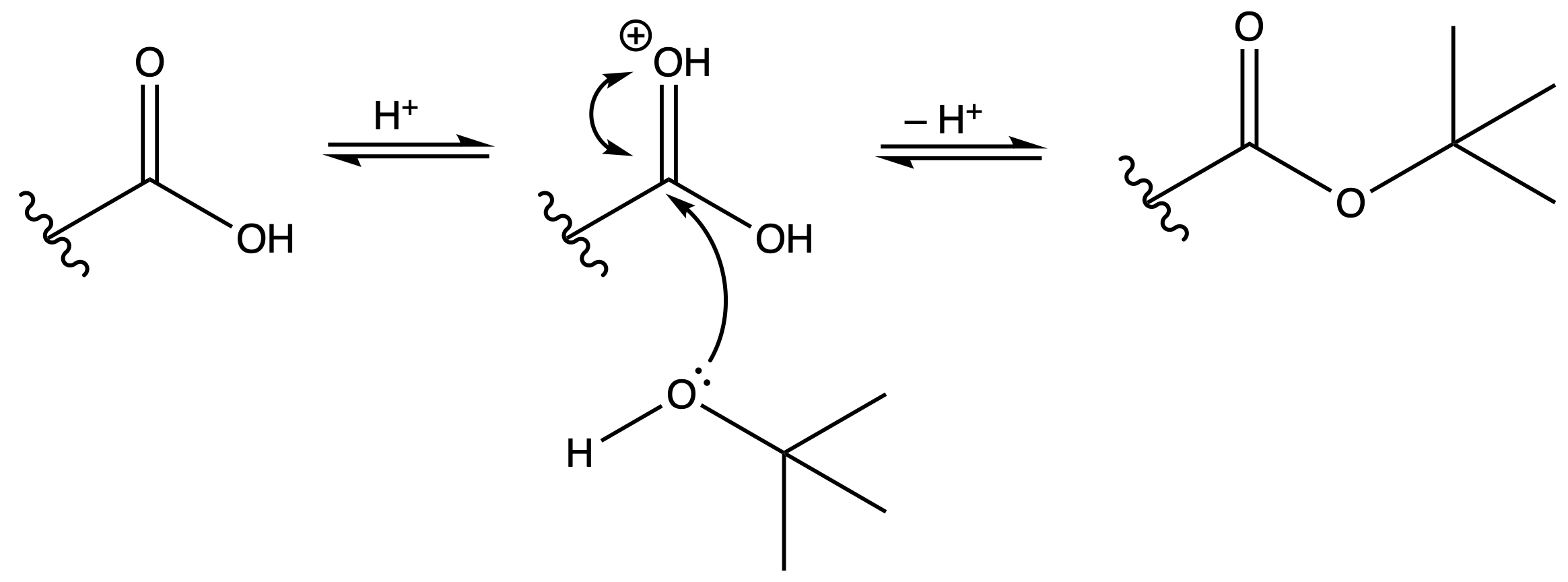
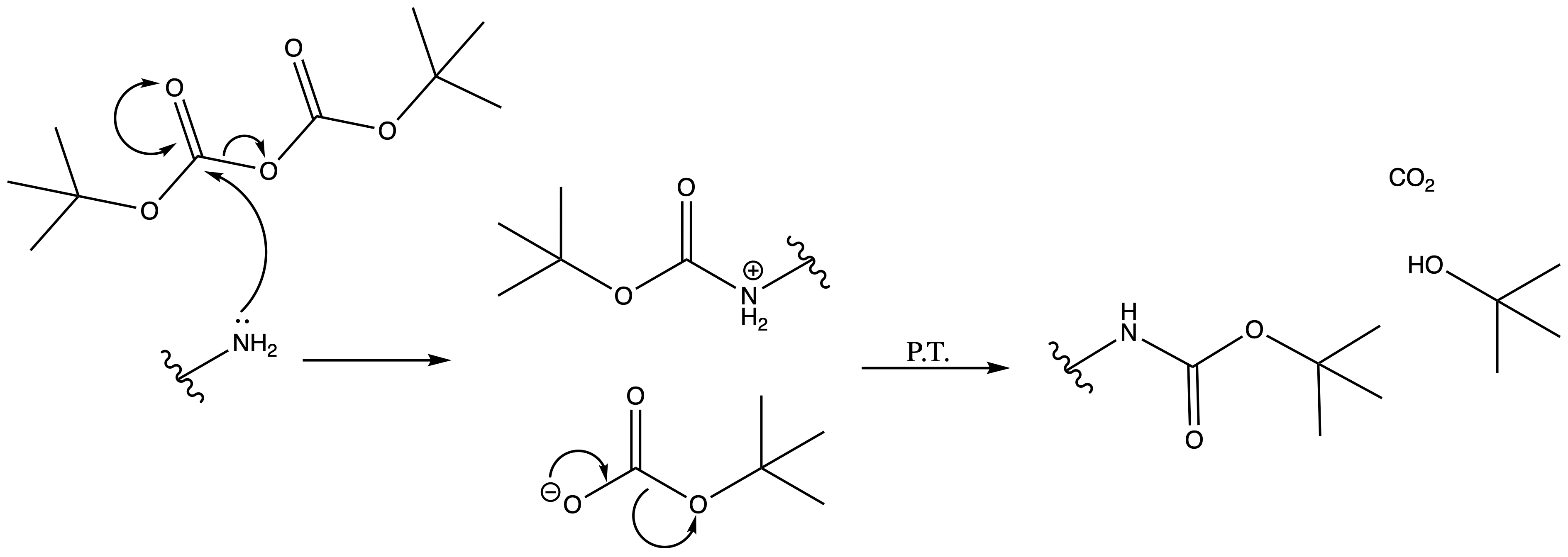
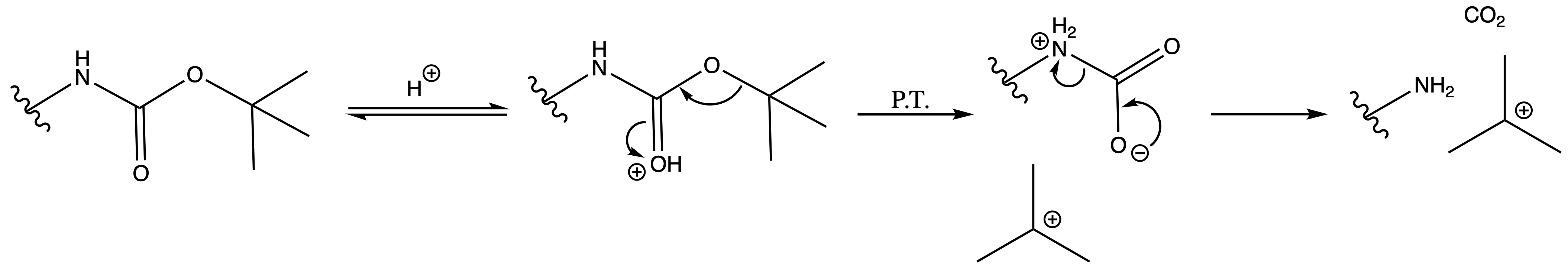
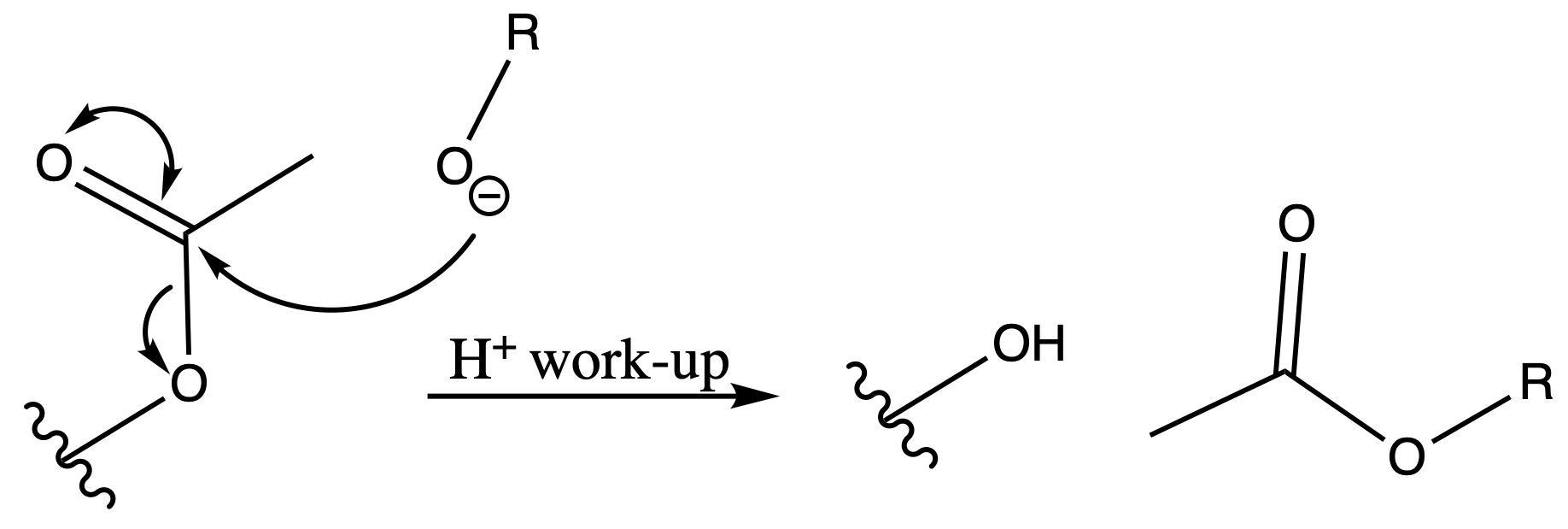
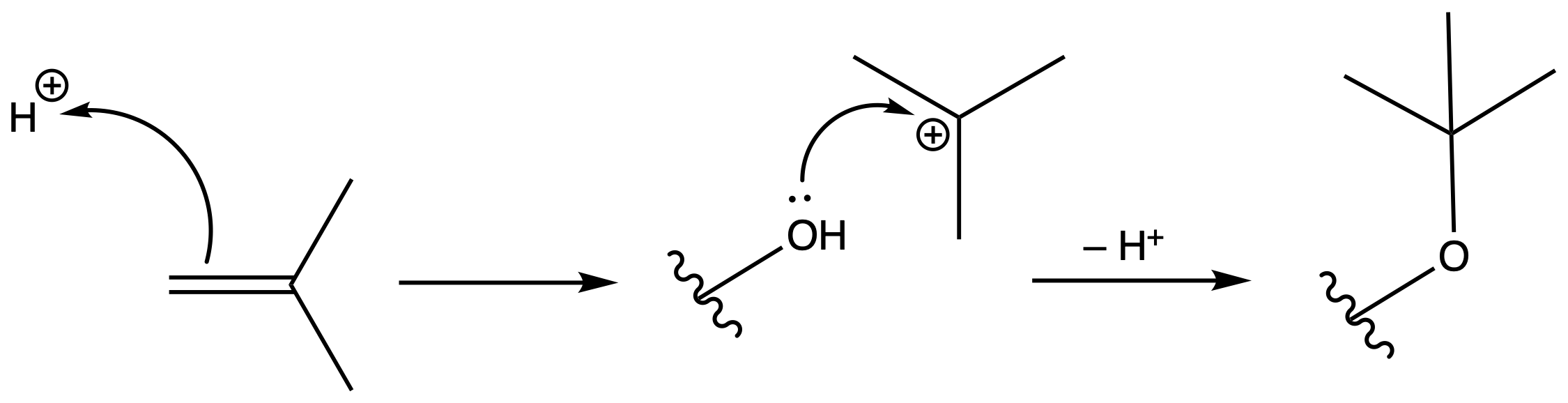
Tertbutyl Esters (tBu)
Protection
Reagents needed:
- tBu–OH + H+
or- tBu–OH + DCC
Mechanism :
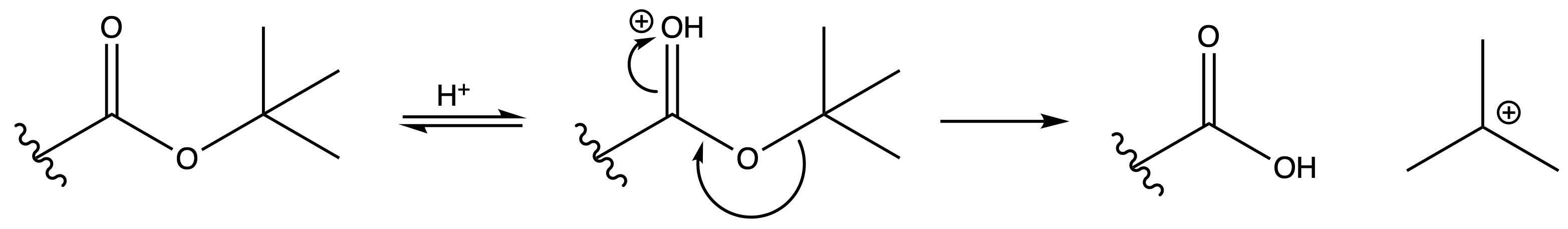
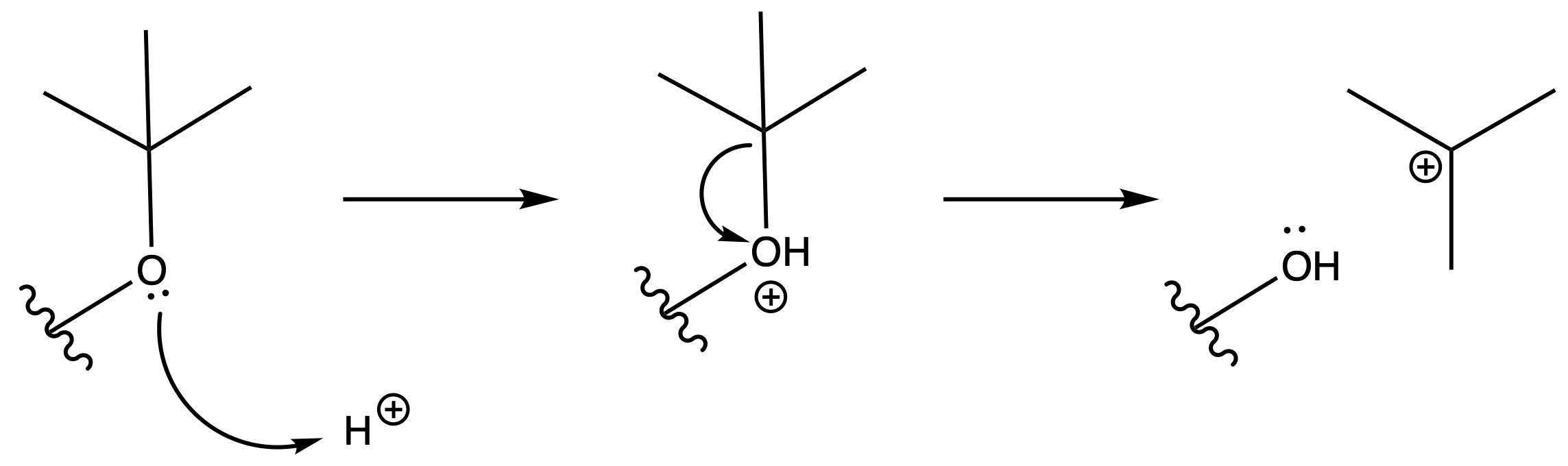
Deprotection
Reagents needed:
- Concentrated acid
Mechanism :
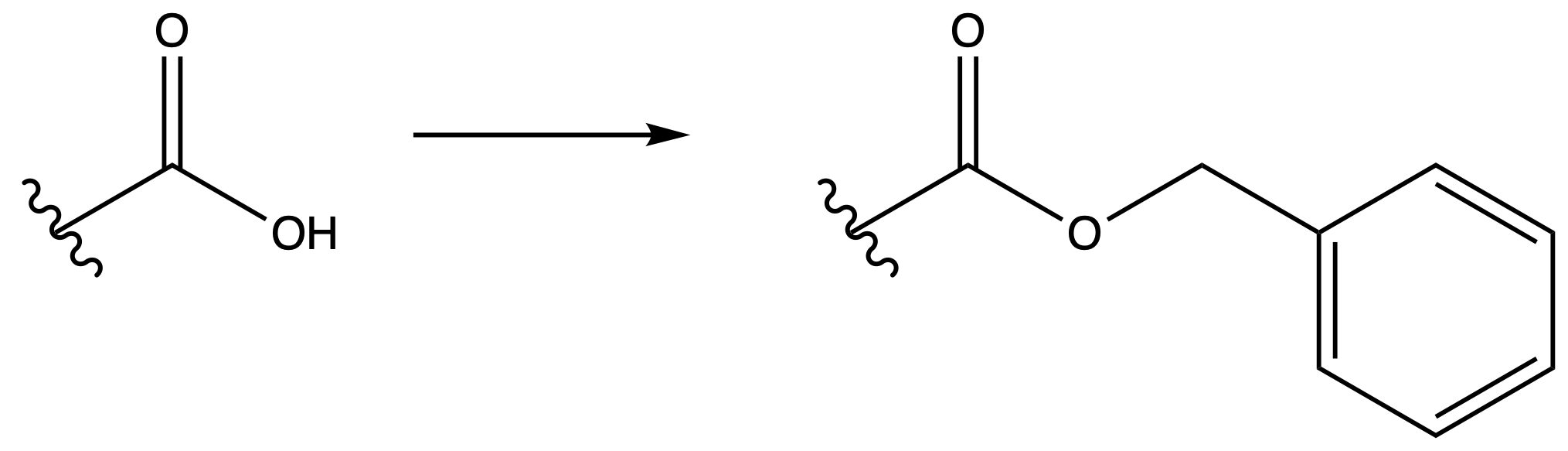
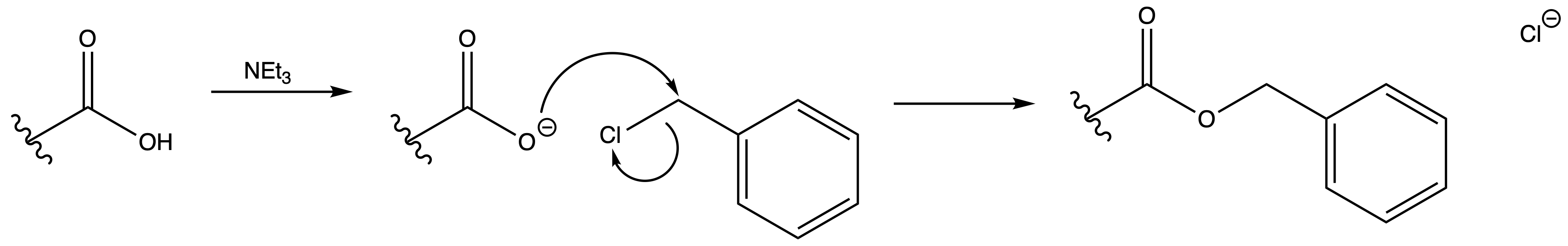
Benzyl ester (Bn)
ProtectionReagents needed:
- Bn-Cl + NEt3
Mechanism :
Deprotection
Reagents needed:
- H2 + Pd/C
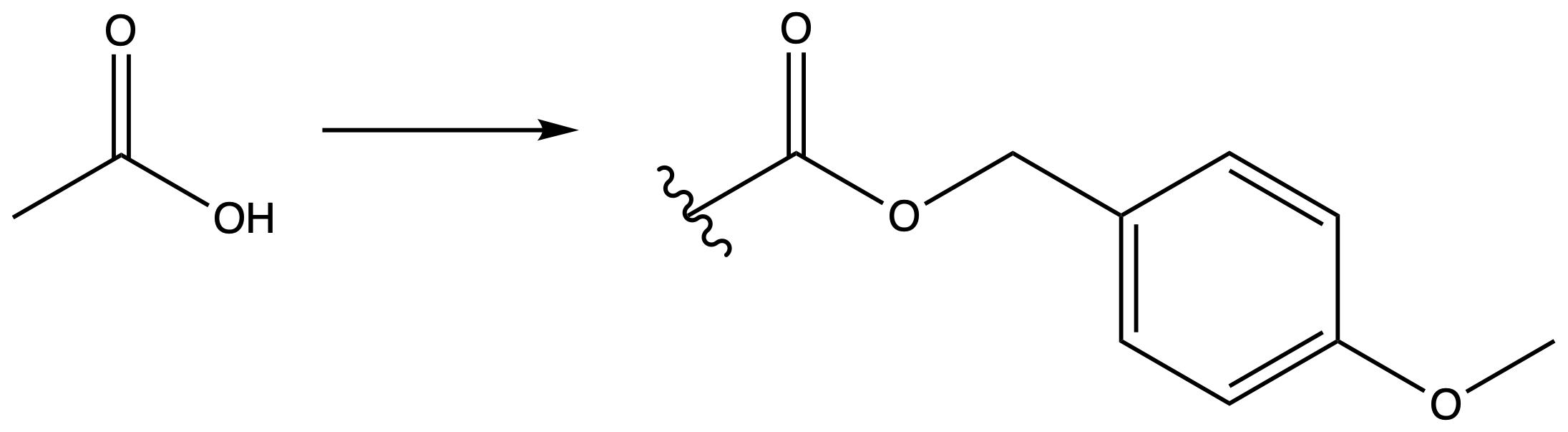
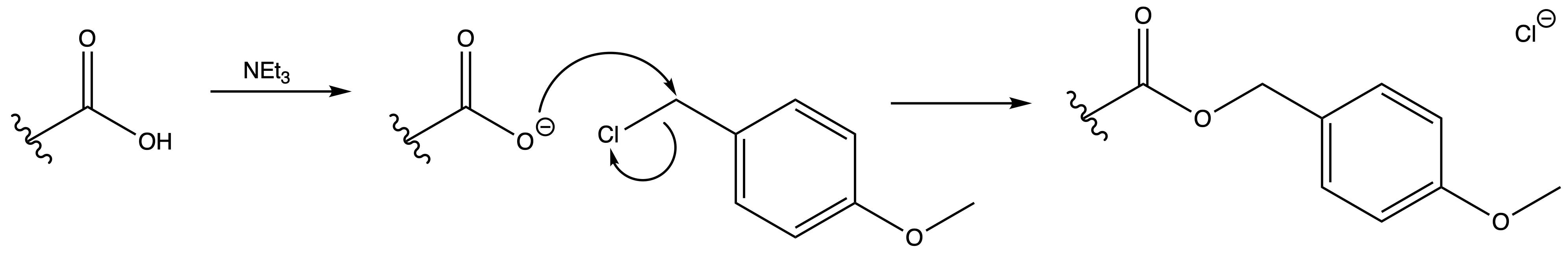
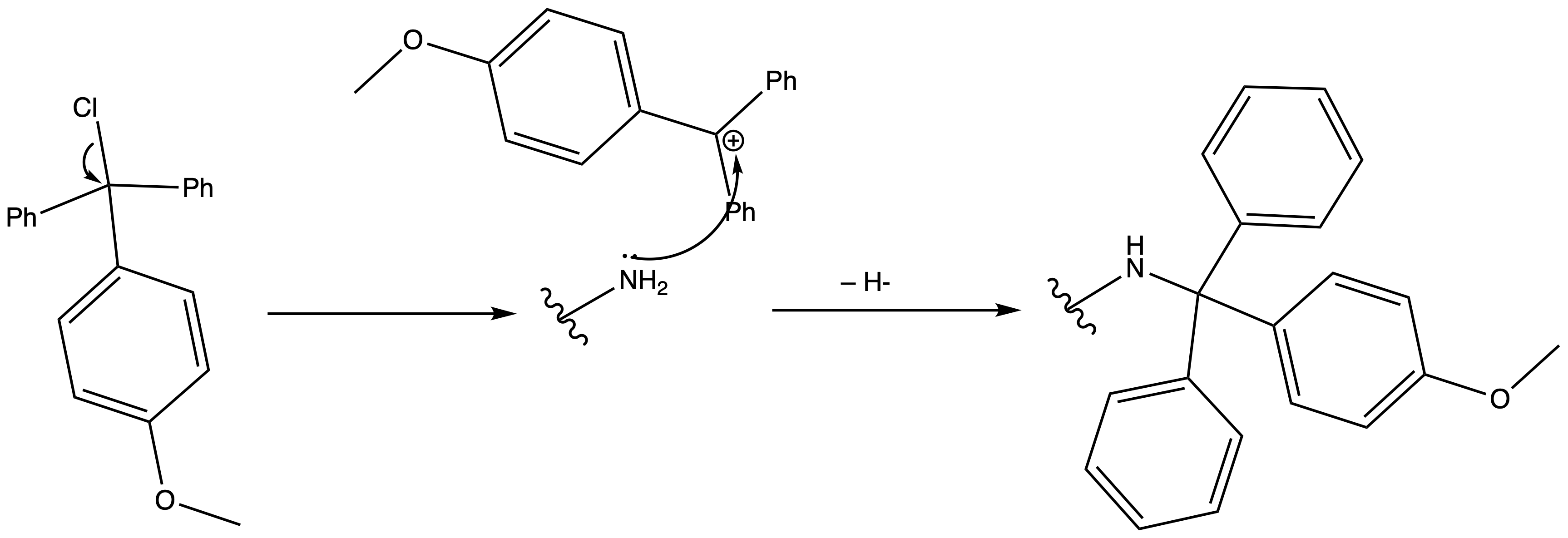
4-Methoxybenzyl Ester (PMB)
ProtectionReagents needed:
- PMB-Cl + NEt3
Mechanism :
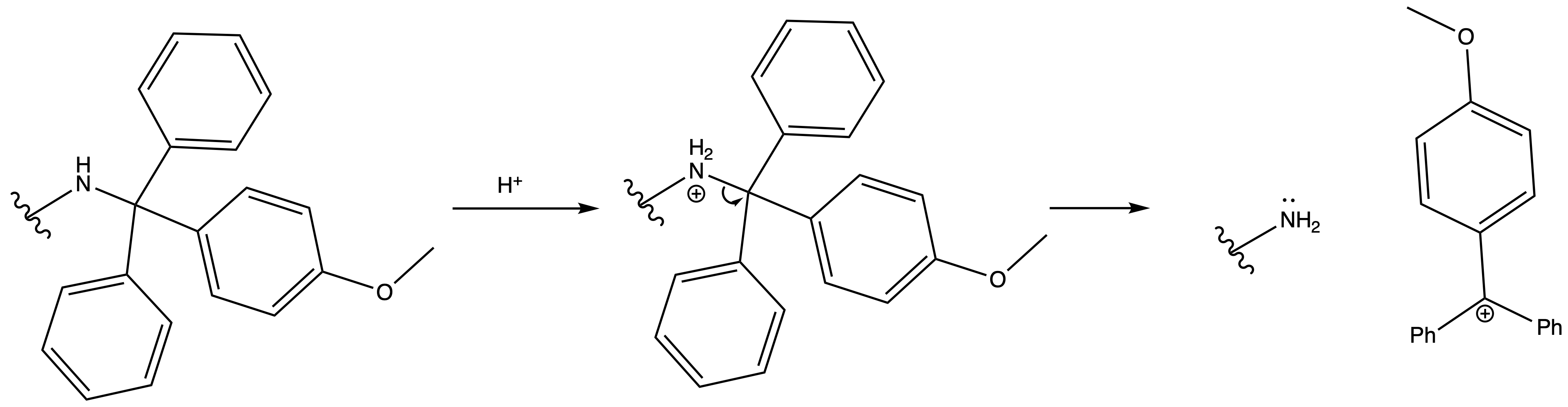
Deprotection
Reagents needed:
- Concentrated acid
or- H2 + Pd/C
¶ Amines
We want to protect the amine group so as to
- stop the nitrogen from acting as a nucleophile
- stop the nitrogen from acting as a base
We can achieve this by converting the amine group into a carbamates group such that the steric hinderance and conjugation will kinetically and thermodynamically prevent reactions
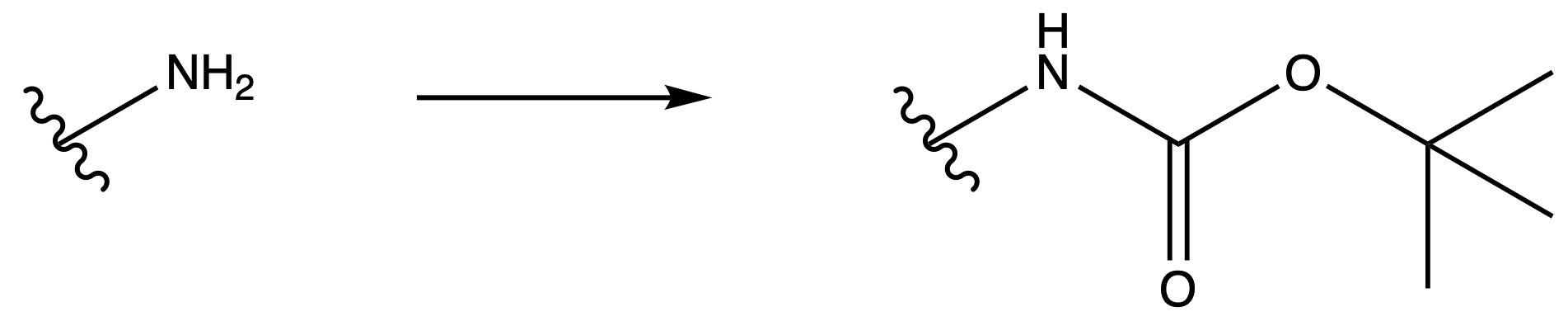
tert-Butyloxycarbonyl Carbamate ( Boc )
ProtectionReagents needed:
- Boc2O + DMAP
Mechanism :
Deprotection
Reagents needed:
- Concentrated acid
Mechanism :
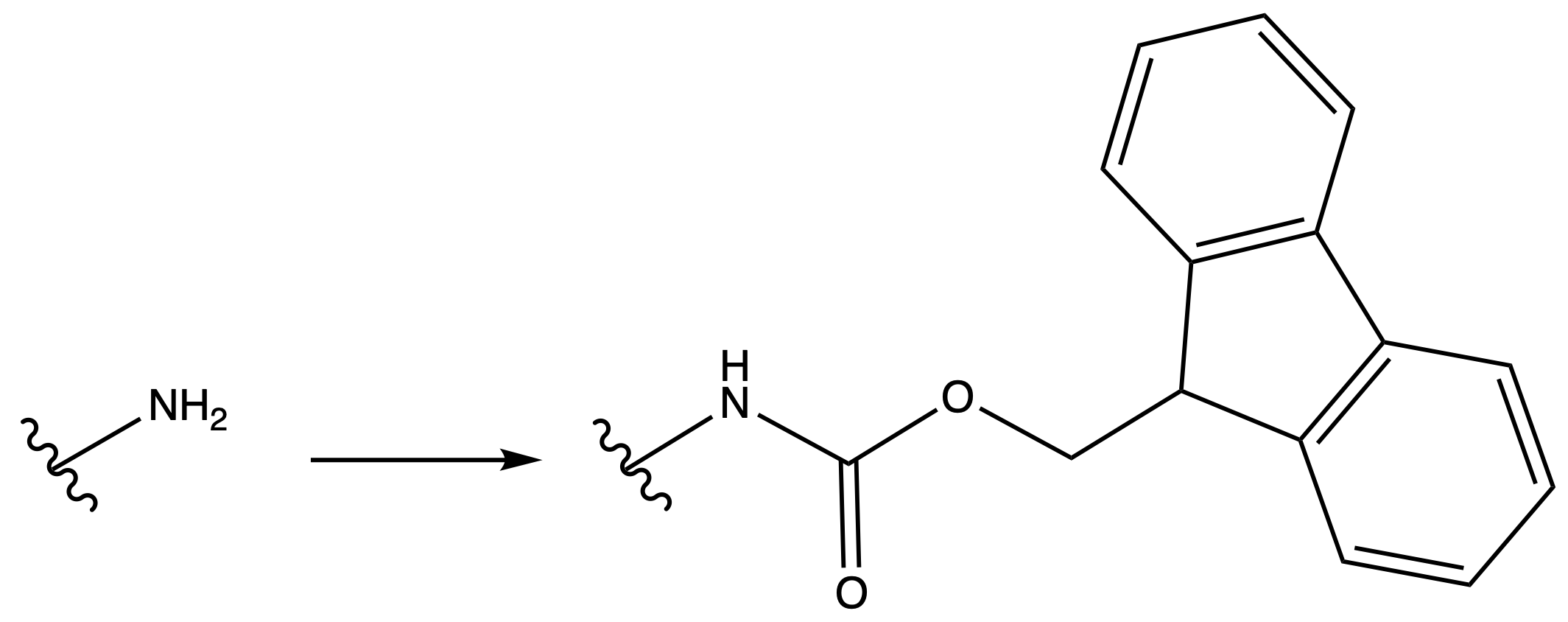
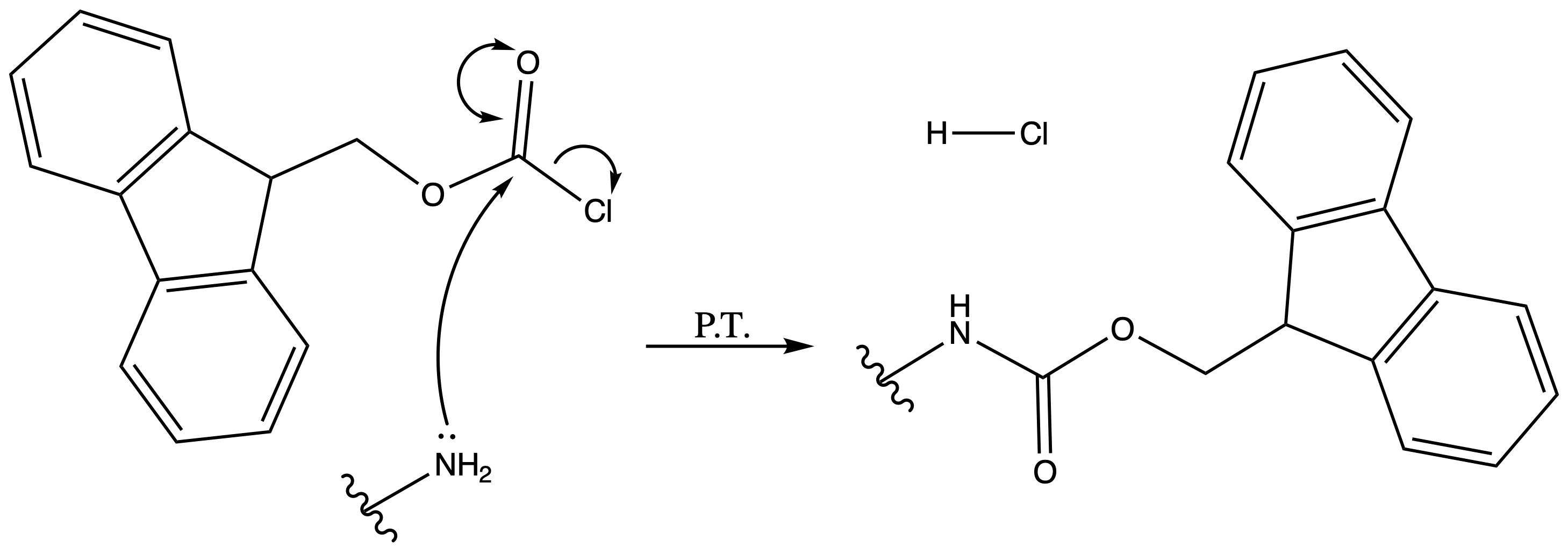
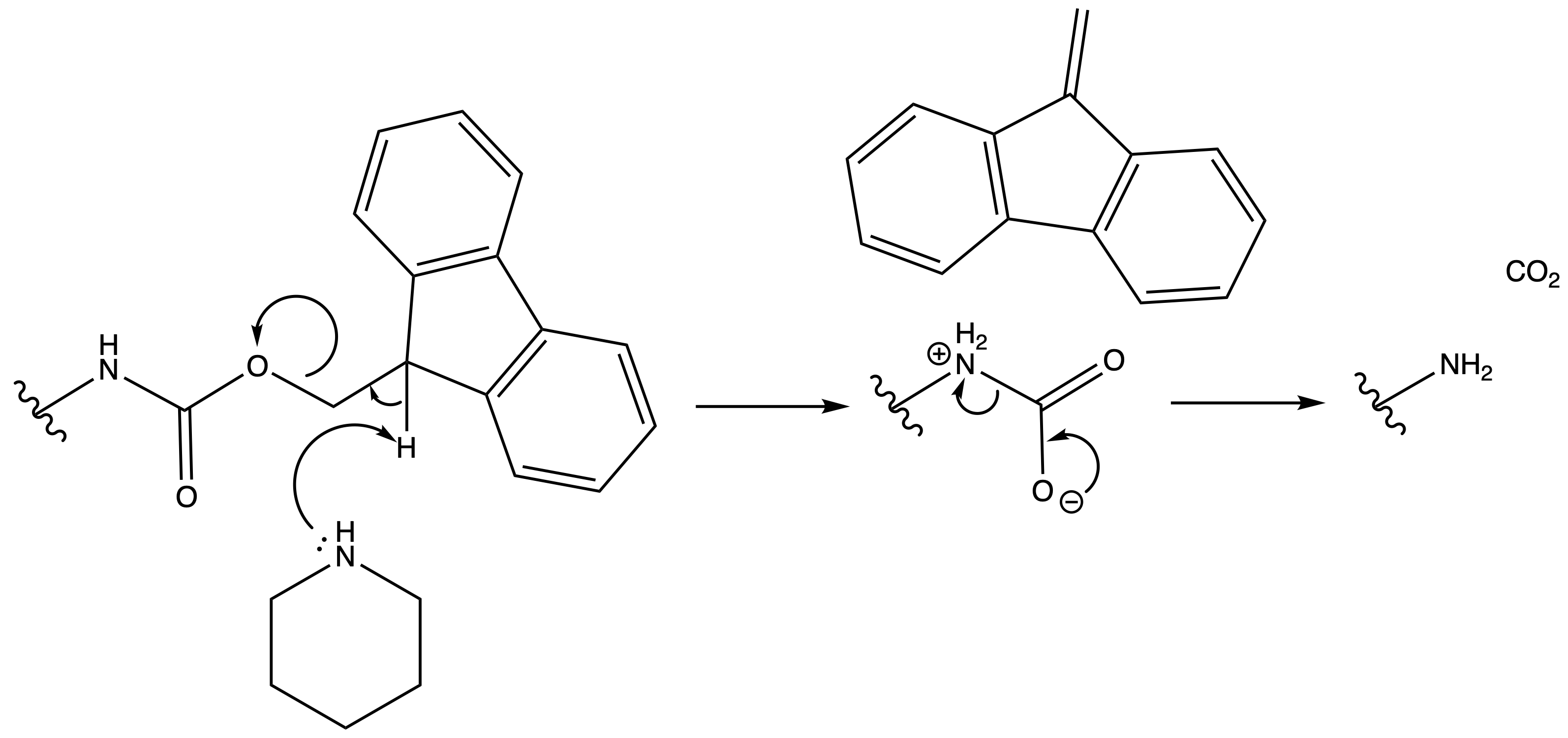
9-Fluorenylmethoxycarbonyl Carbamate ( Fmoc )
ProtectionReagents needed:
- Fmoc-Cl
Mechanism :
Deprotection
Reagents needed:
- Base
Mechanism :
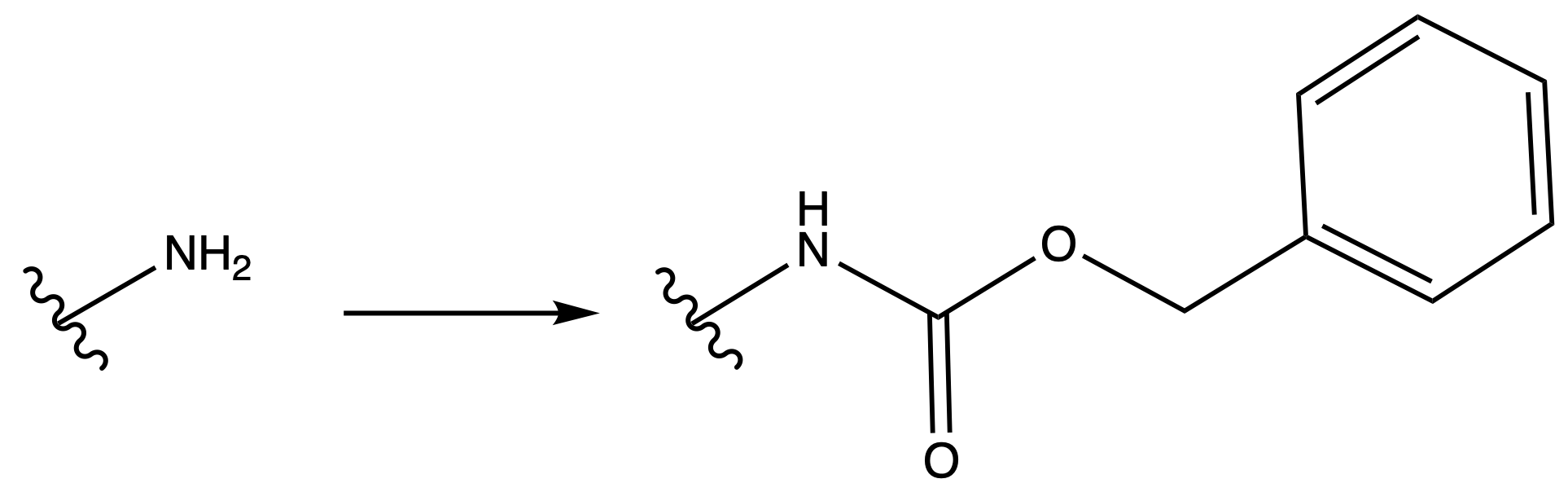
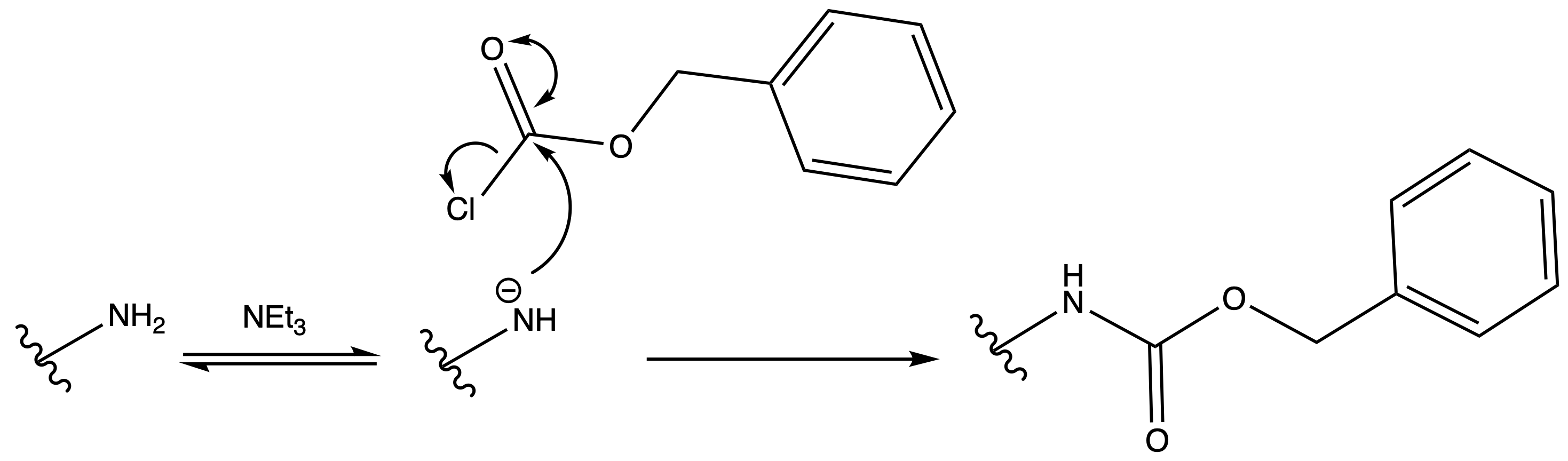
Carboxybenzyl Carbamate ( Cbz )
ProtectionReagents needed:
- Boc2O + DMAP
Mechanism :
Deprotection
Reagents needed:
- H2 + Pd/C
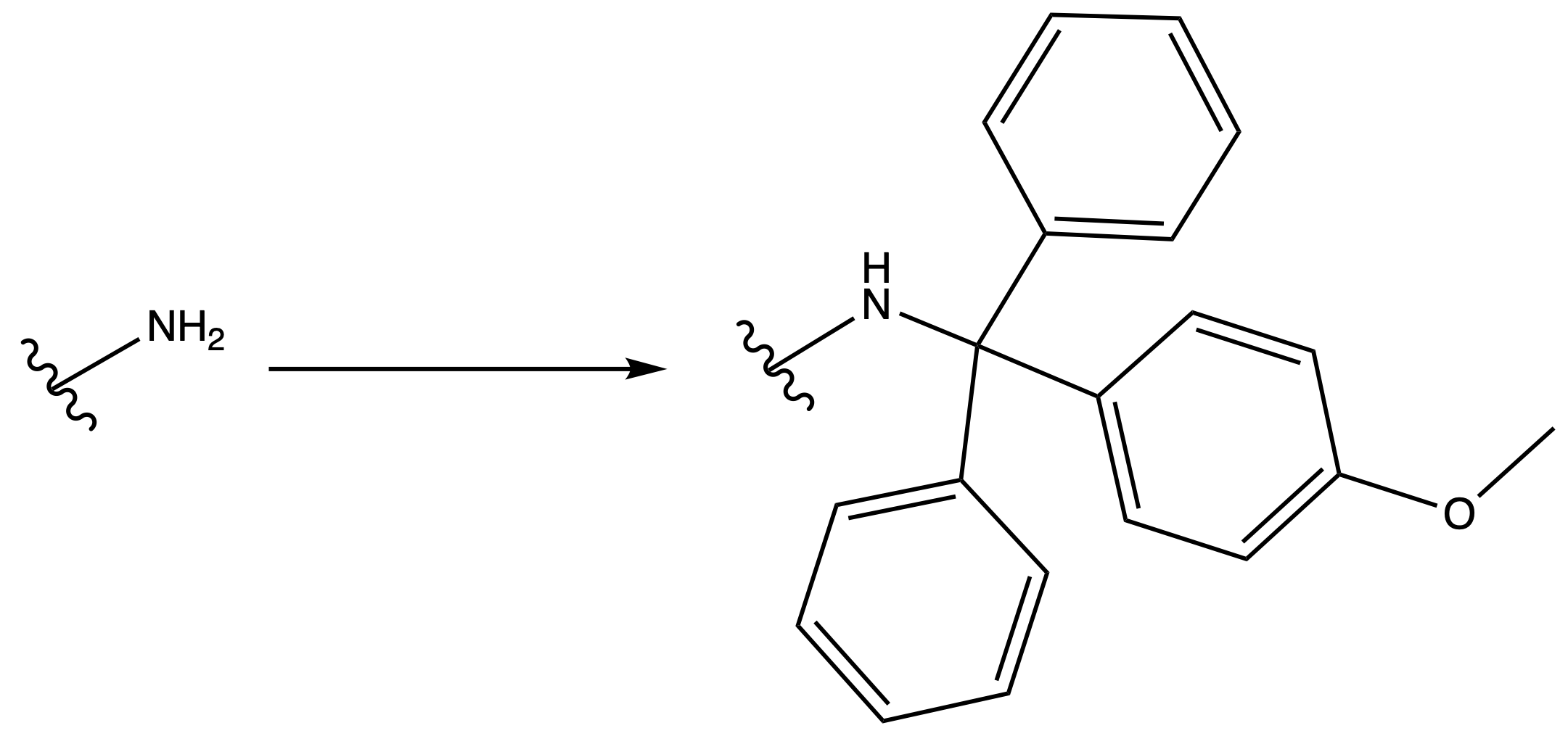
Monomethoxytrityl ( MMT )
ProtectionReagents needed:
- MMT–Cl
Mechanism :
Deprotection
Reagents needed:
- Concentrated acid
Mechanism :
¶ Alochol
We want to protect the amine group so as to
- stop the oxygen acting as a nucleophile,
- prevent oxidation
- proton donation
We can achieve this by introducing steric hindrance or conjugation
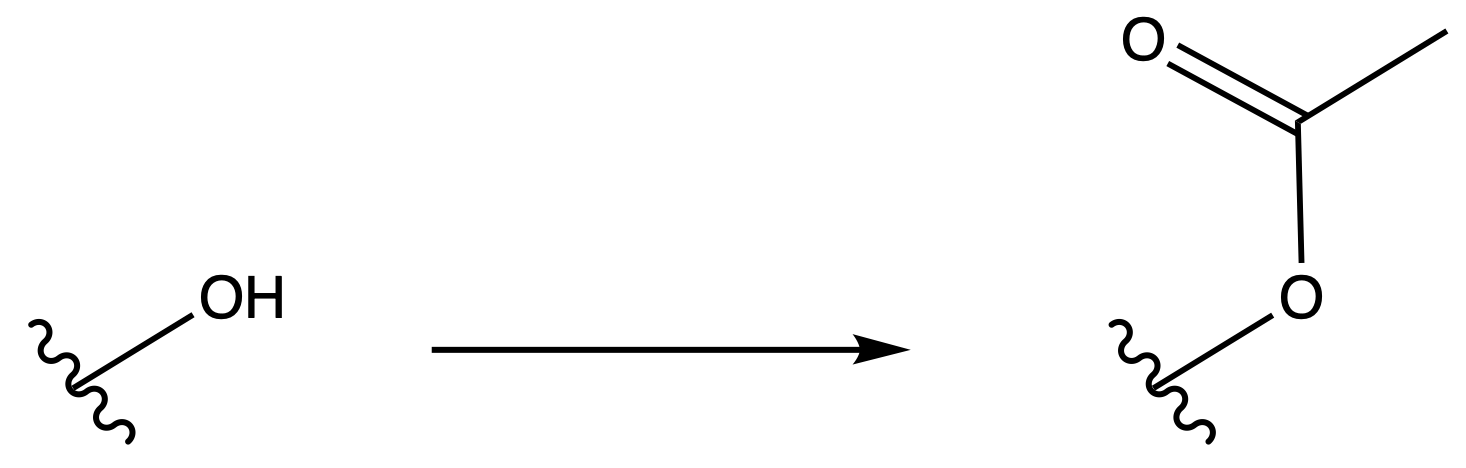
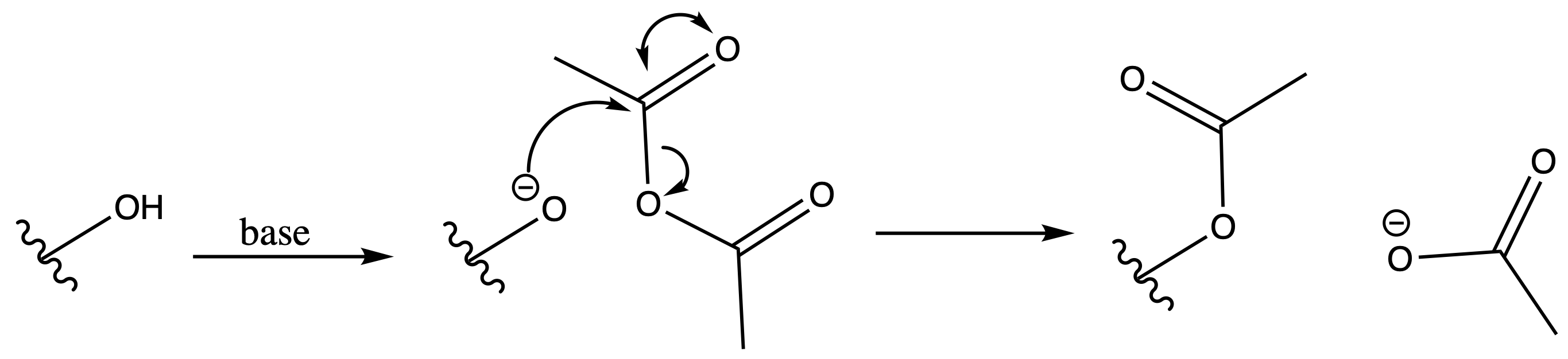
Ester
ProtectionReagents needed:
- Ac2O + Base
Mechanism :
Deprotection
Reagents needed:
- Strong Base
Mechanism :
¶
tert-butyl Eter
ProtectionReagents needed:
- Ac2O + Base
Mechanism :
Deprotection
Reagents needed:
- Concentrated Acid
Mechanism :
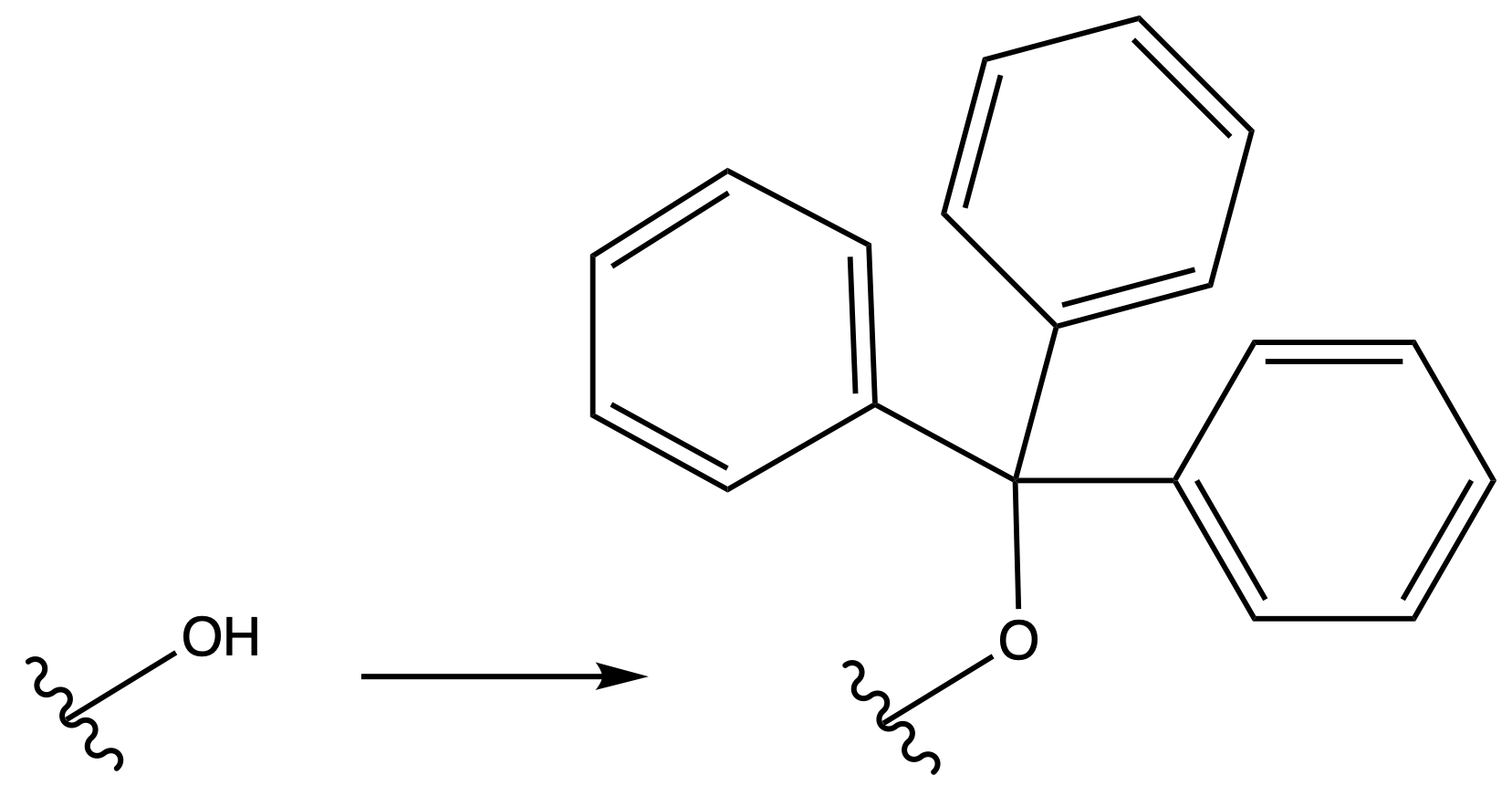
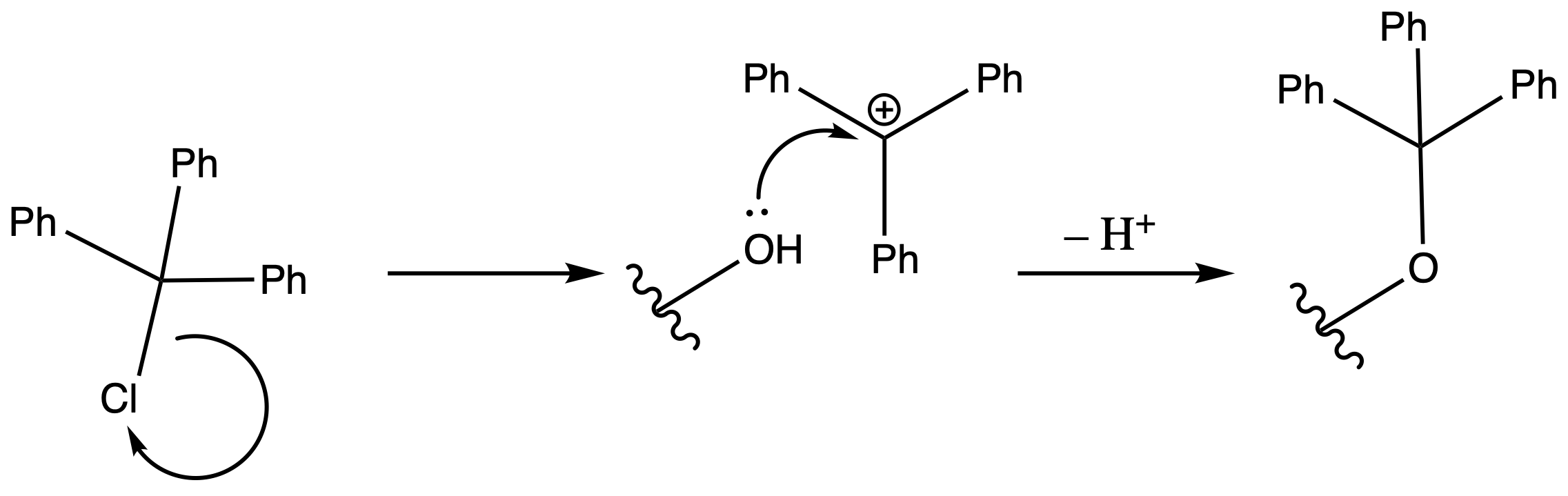
Triphenyl Eter
ProtectionReagents needed:
- Trt–Cl
Mechanism :
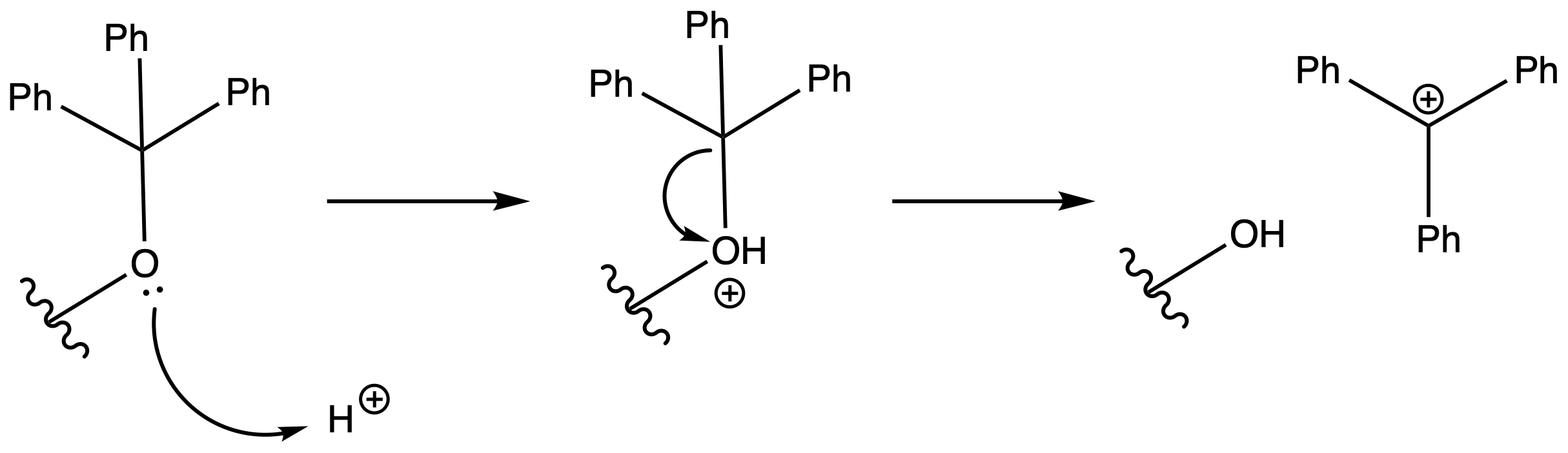
Deprotection
Reagents needed:
- Dilute Acid
Mechanism :
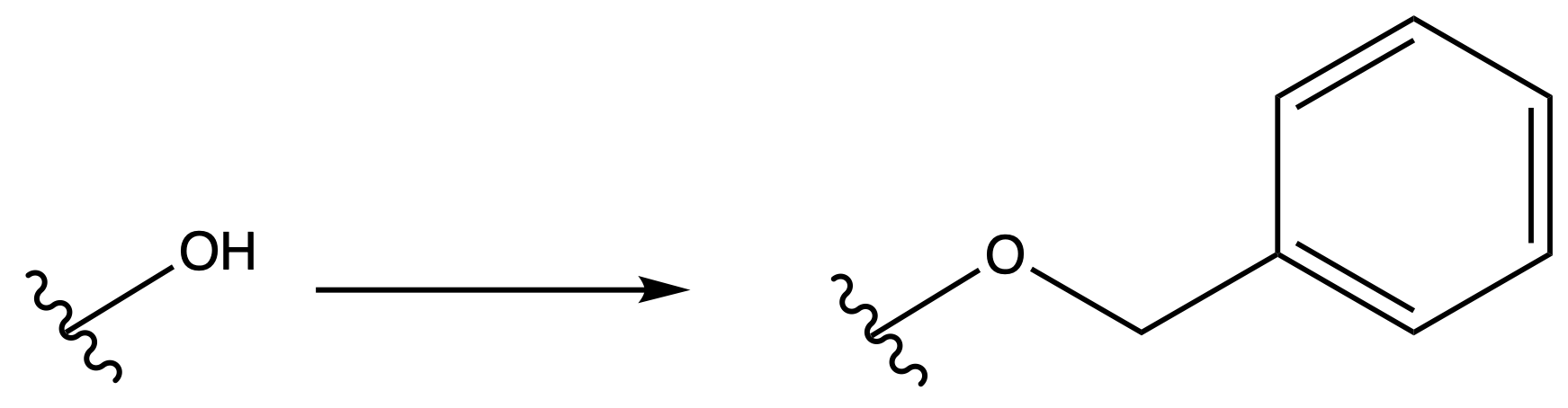
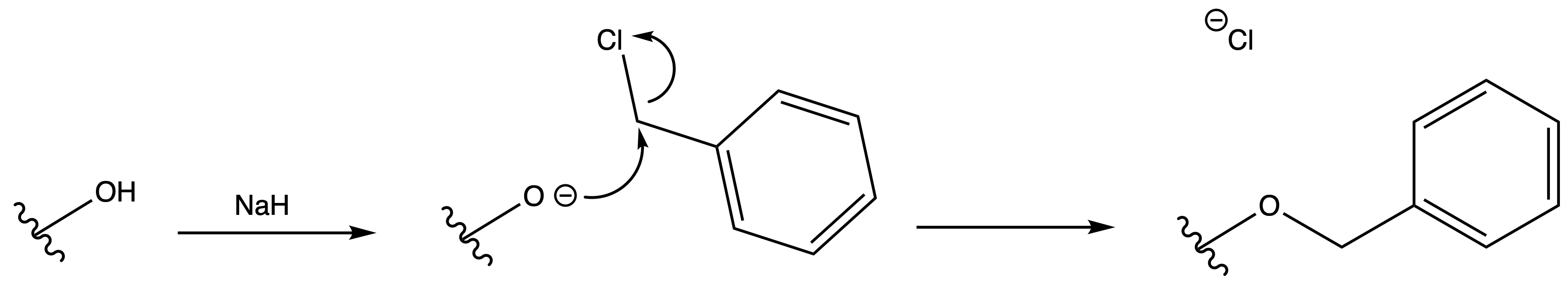
Benzyl Eter ( Bn )
ProtectionReagents needed:
- Bn–Cl + NaH
Mechanism :
Deprotection
Reagents needed:
- H2 + Pd/C
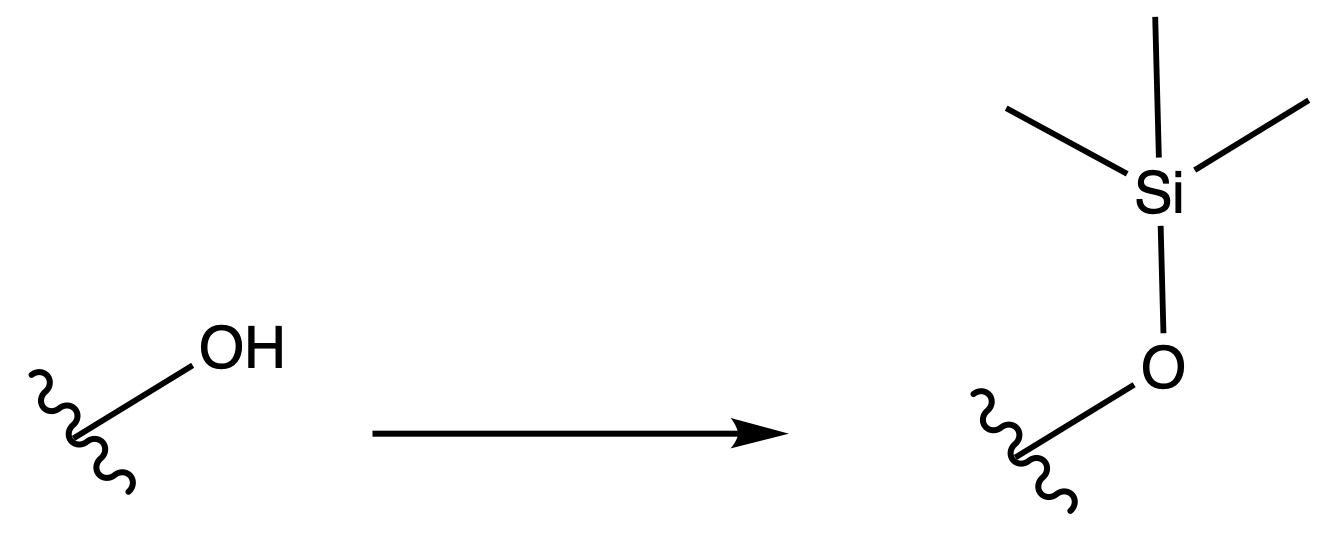
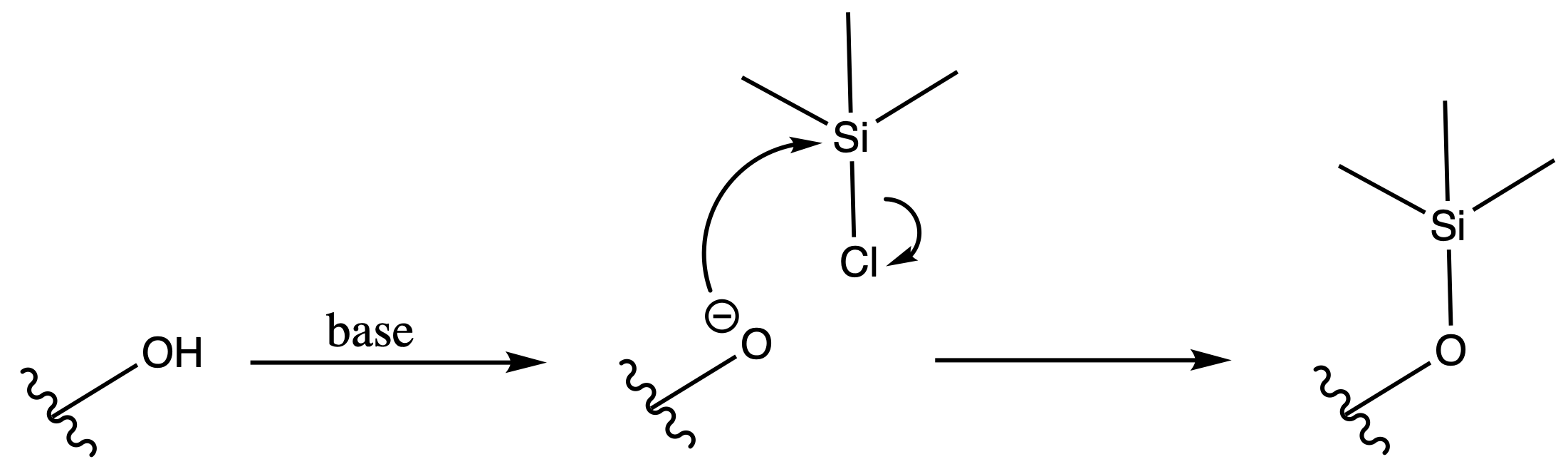
Trimethylsilyl Eter ( TMS )
ProtectionReagents needed:
- TMSCl
Mechanism :
Deprotection
Reagents needed:
- Concentrated Acid
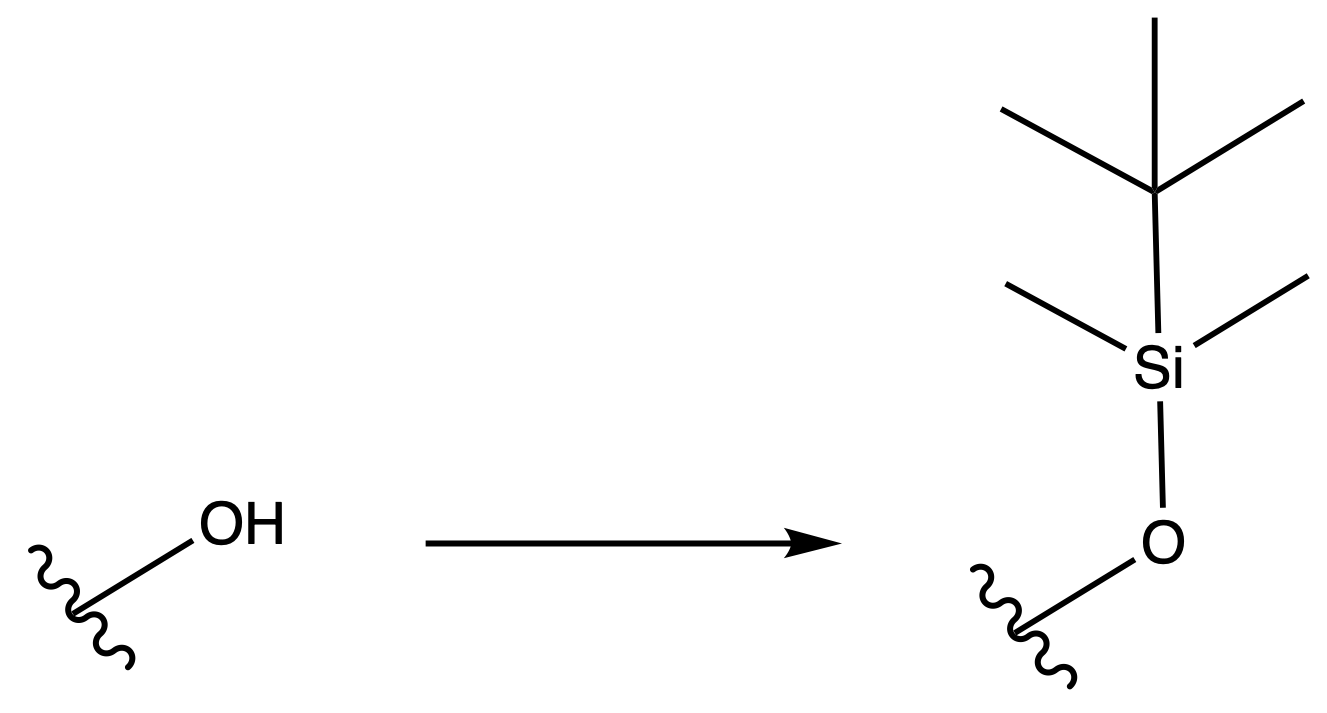
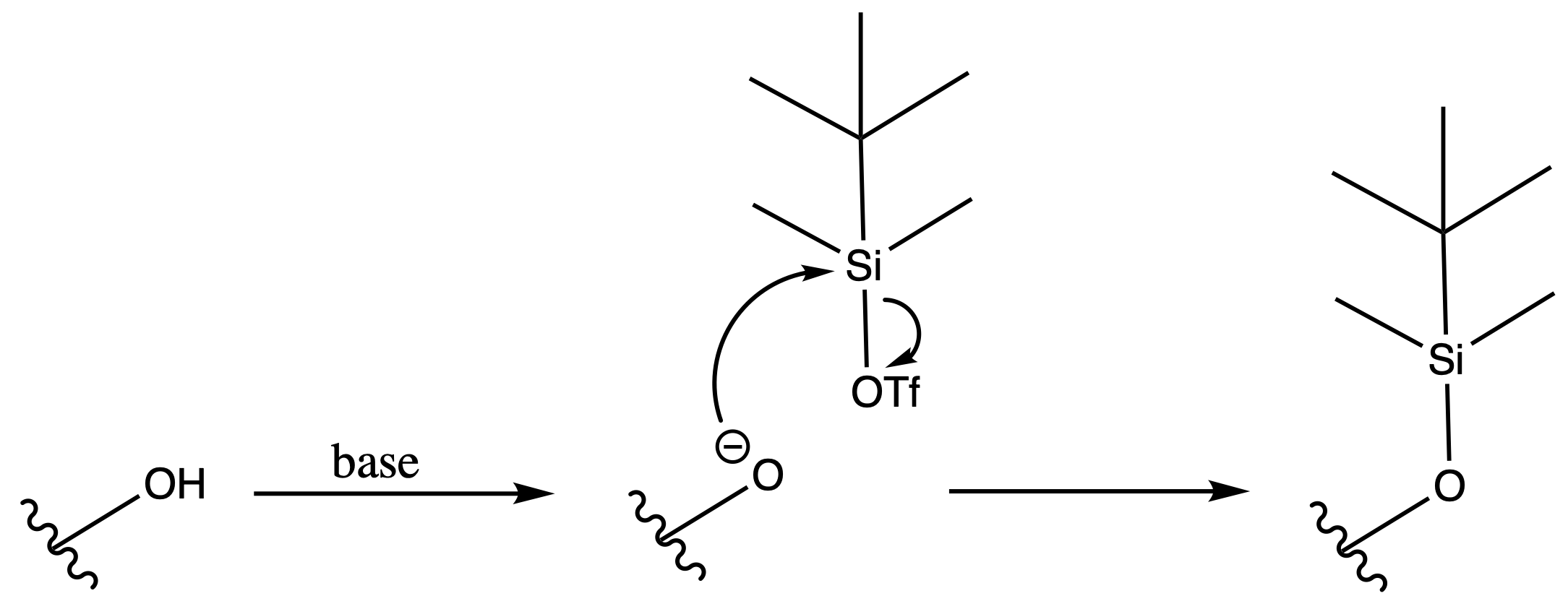
Tert-butyldimethylsilyl Eter ( TBDMS )
ProtectionReagents needed:
- TBDMS-OTf
Mechanism :
Deprotection
Reagents needed:
- Concentrated Acid
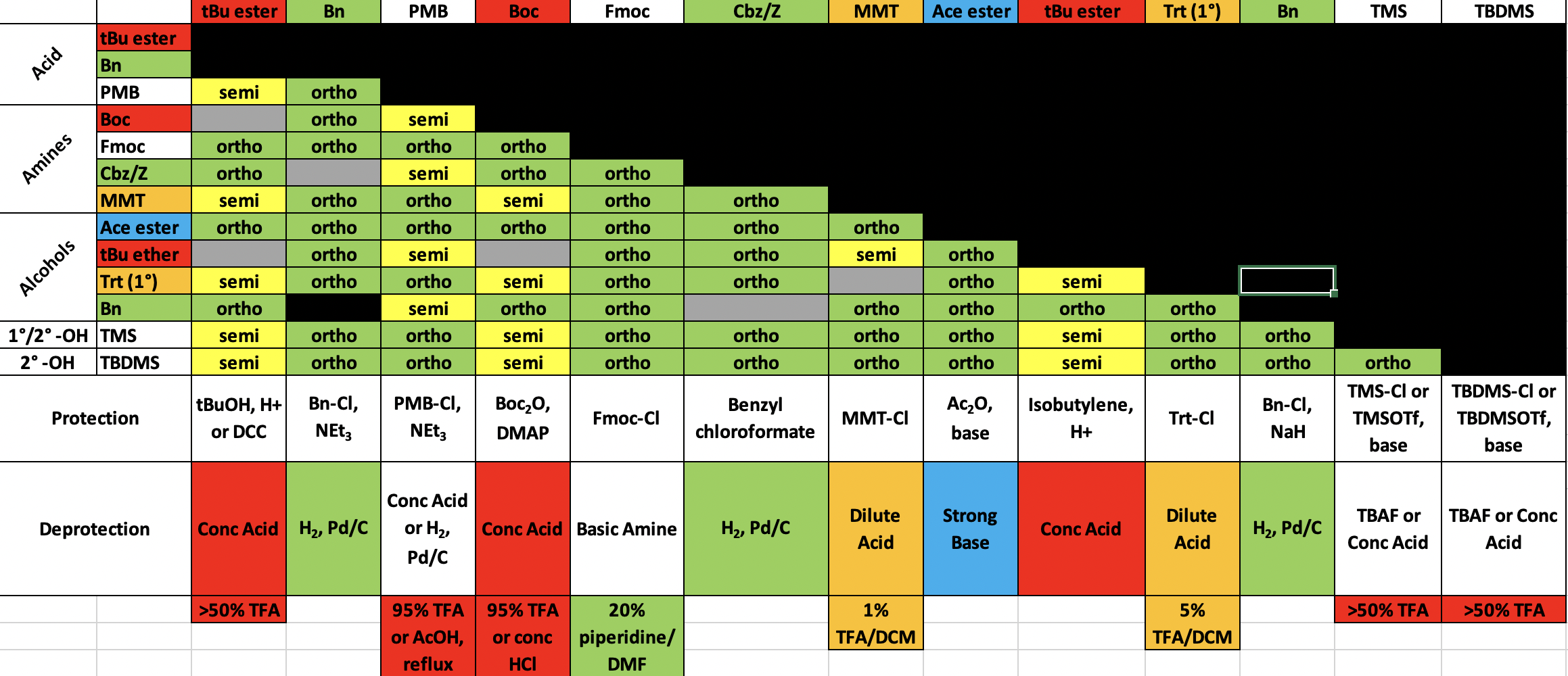
¶ Summary
¶ Amino Acid and Peptides
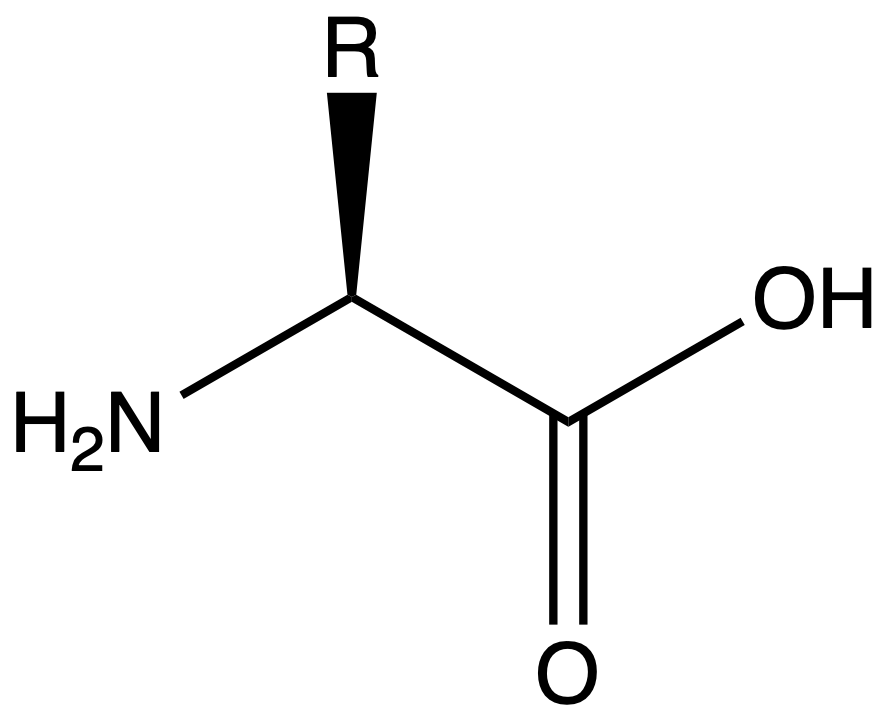
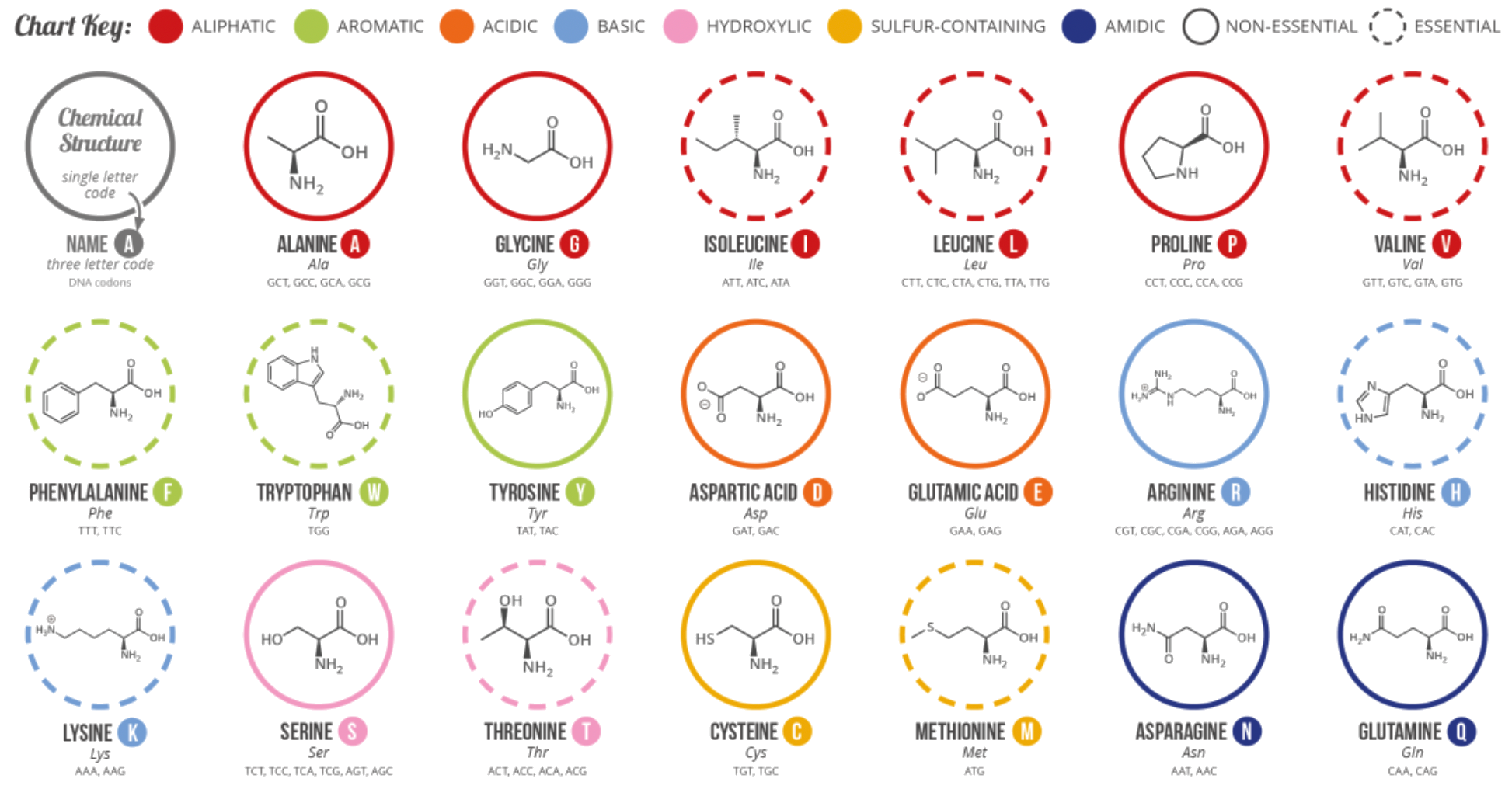
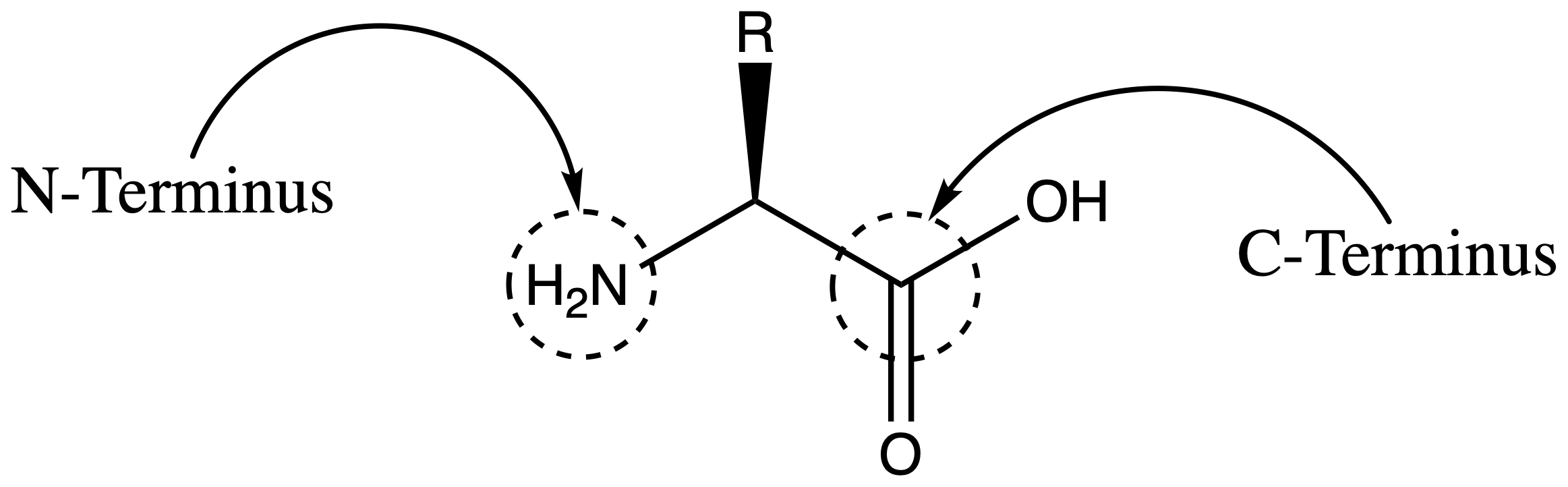
Natural amino acid is a molecule with a general structure as such
- The amine group is to the carboxyl group
- The alpha carbon is either a chiral or a prochiral centre
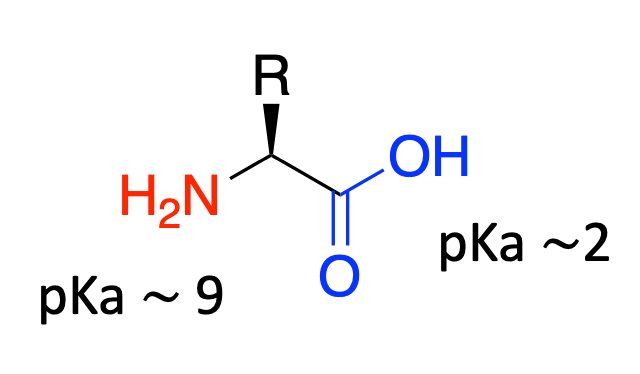
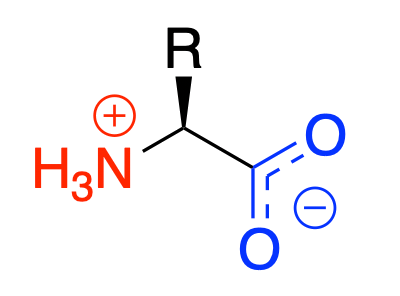
Zwitterionic
Amino acids are zwitterionic in water
- At pH 7, the amine group will be protonated and the carboxyl group will be deprotonated
Types of Amino Acids
There are 20 different types of naturally occuring amino acids, their only differences is the group
- Depending on the group, amino acids can have different properties
- We give each of these amino acid a three-letter code and a one-letter code
Peptides
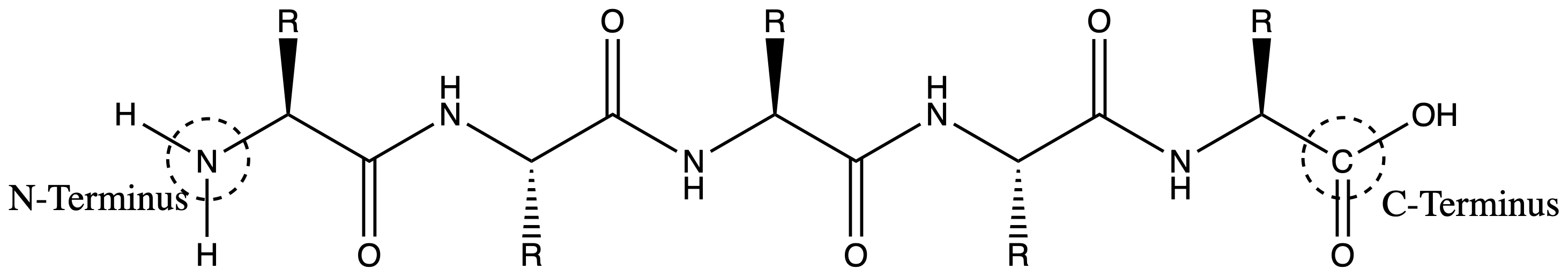
Two amino acids join together via the formation of a peptide bond
- A peptide bond is an amide bond
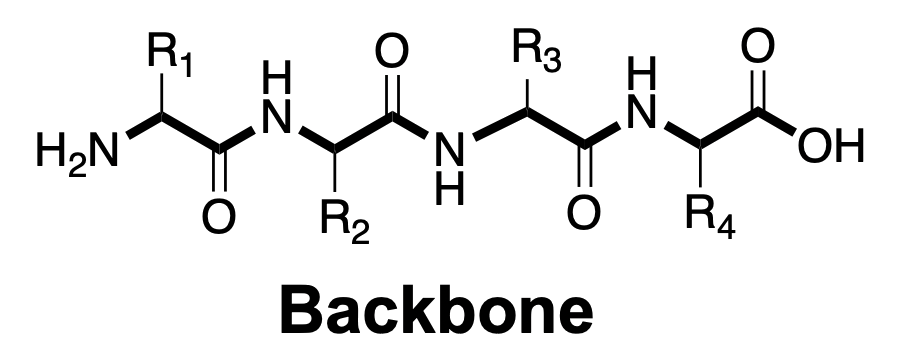
When we want to refer to different part of the peptide, we have two different words
- The backbone describes the extended chain that goes between the amine group and the carboxyl group
- The side chain describes the groups
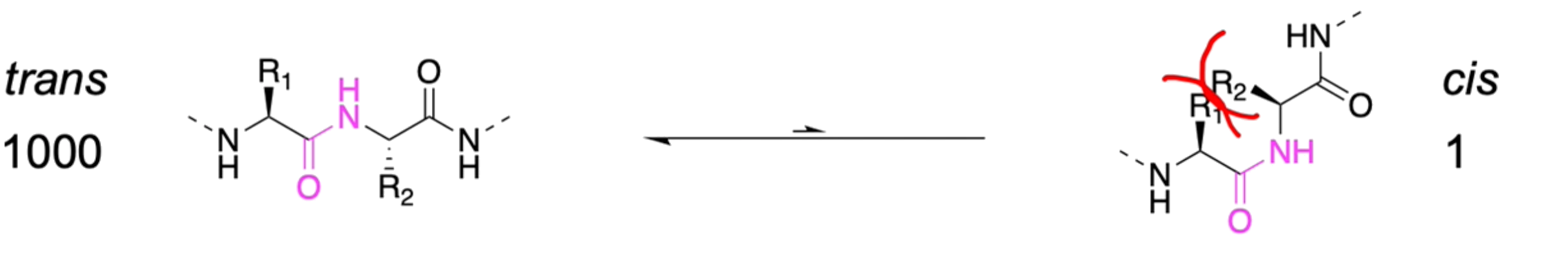
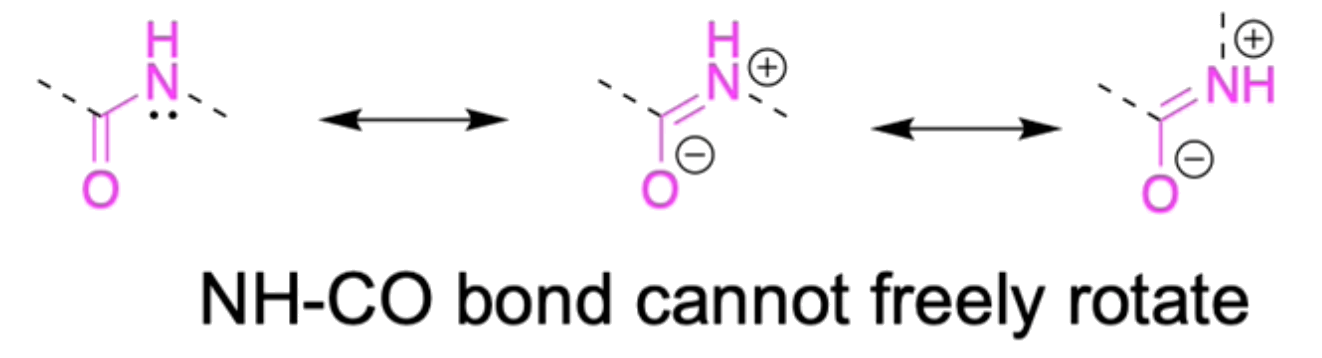
The peptide backbone is in the trans-conformation to reduce steric clashes
- Moreover, the peptide bonds cannot freely rotate since the lone pair on nitrogen is conjugated to the carbonyl, forming a partial double bond
Naming Peptides
An amino acid has two terminal, the N-terminus and the C-terminus
- A peptide therefore also has the N-terminus and the C-terminus
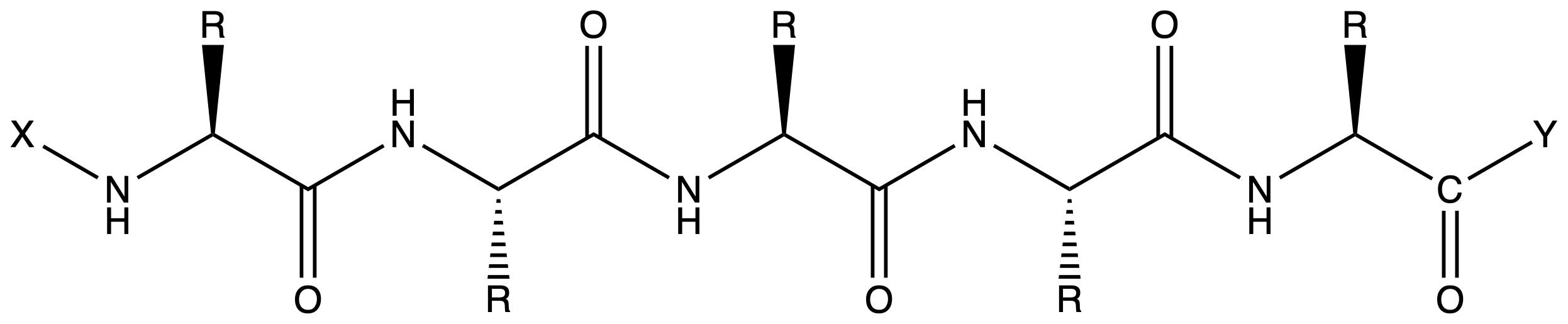
- The N-terminus and the C-terminus can be modified
- We name the peptide from the N-terminus to the C-terminus
¶ Synthetic Strategy of Peptides
Considerations:
- Control over identity of each amino acid
- No racemization
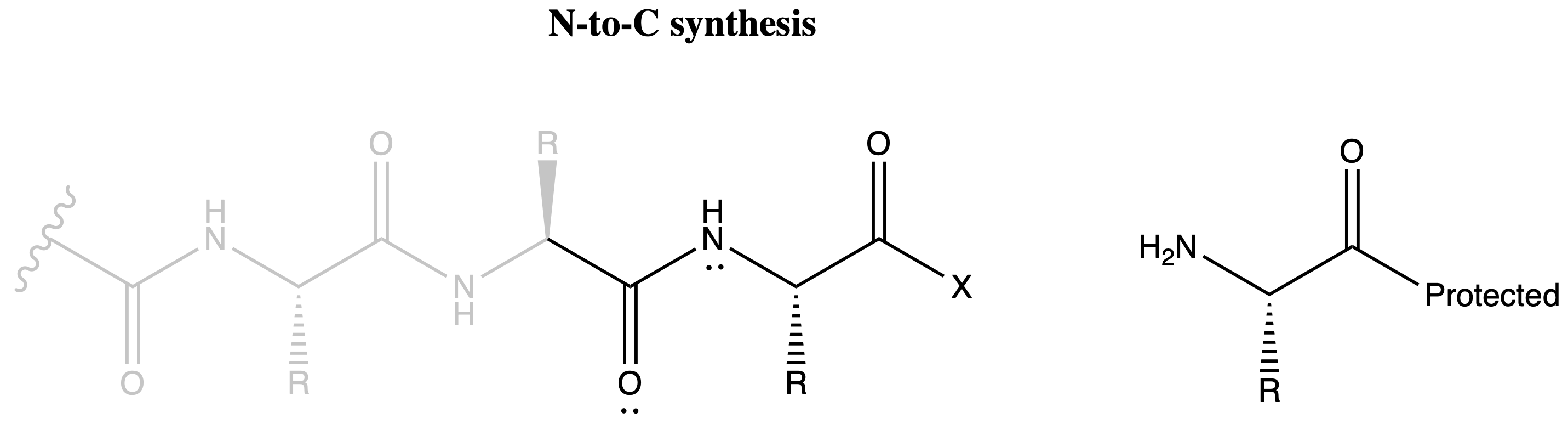
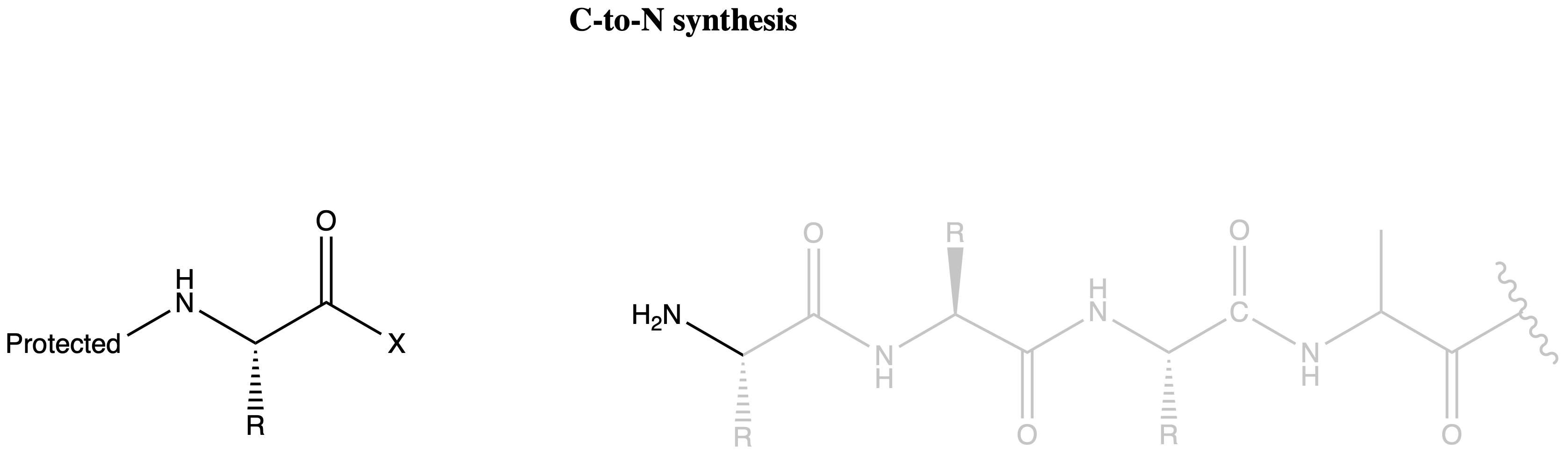
Direction of Peptide Synthesis
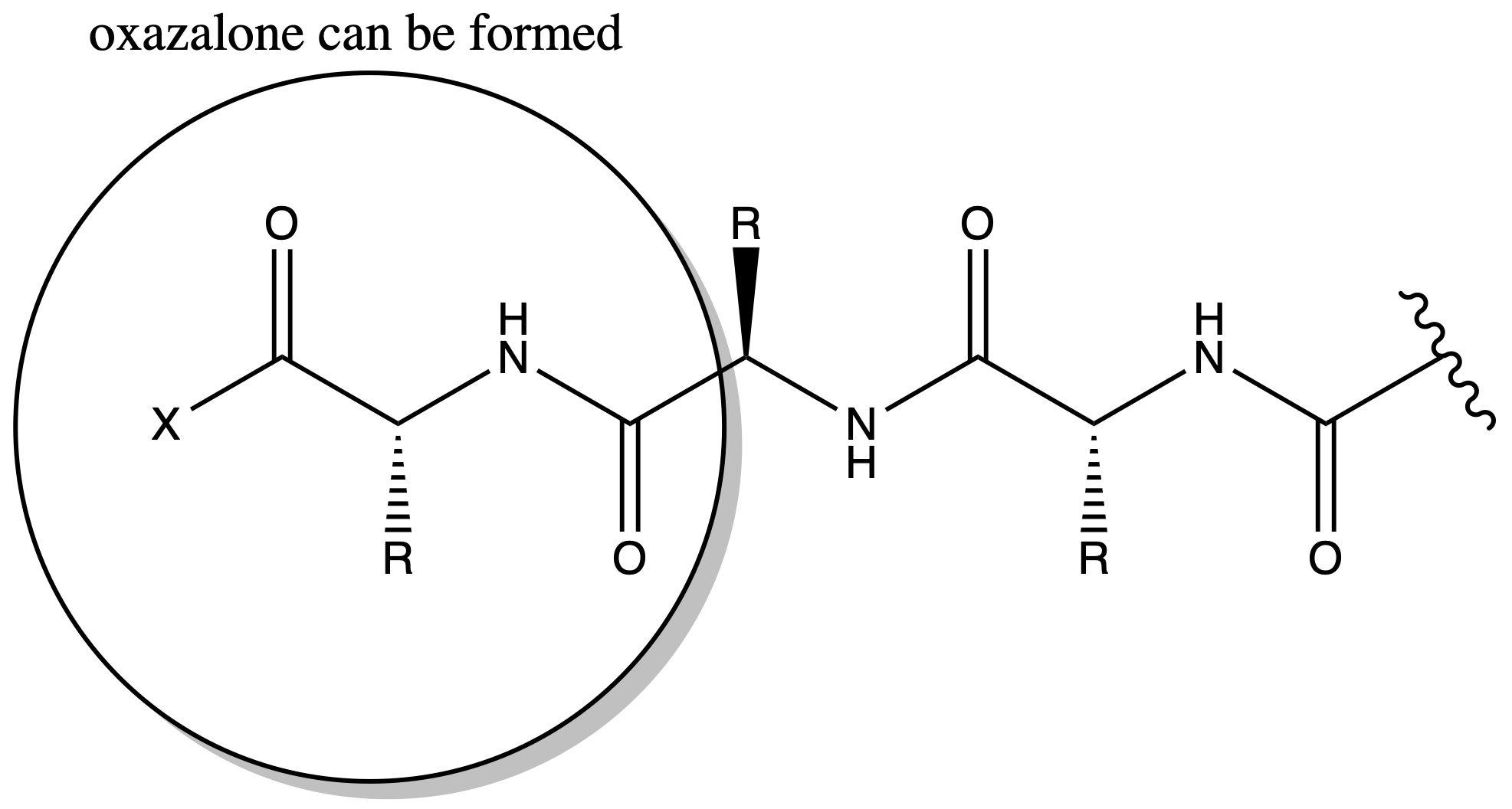
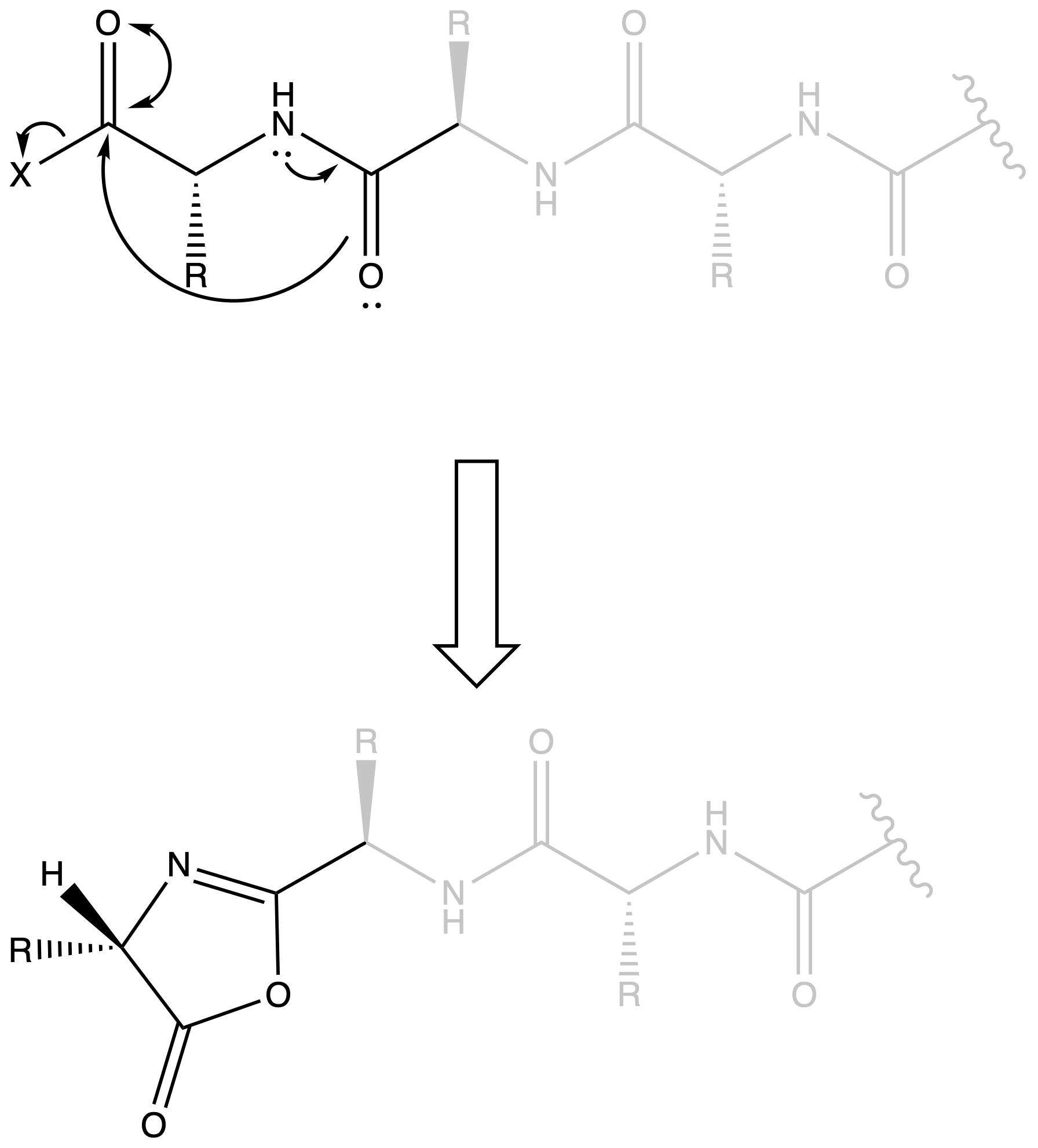
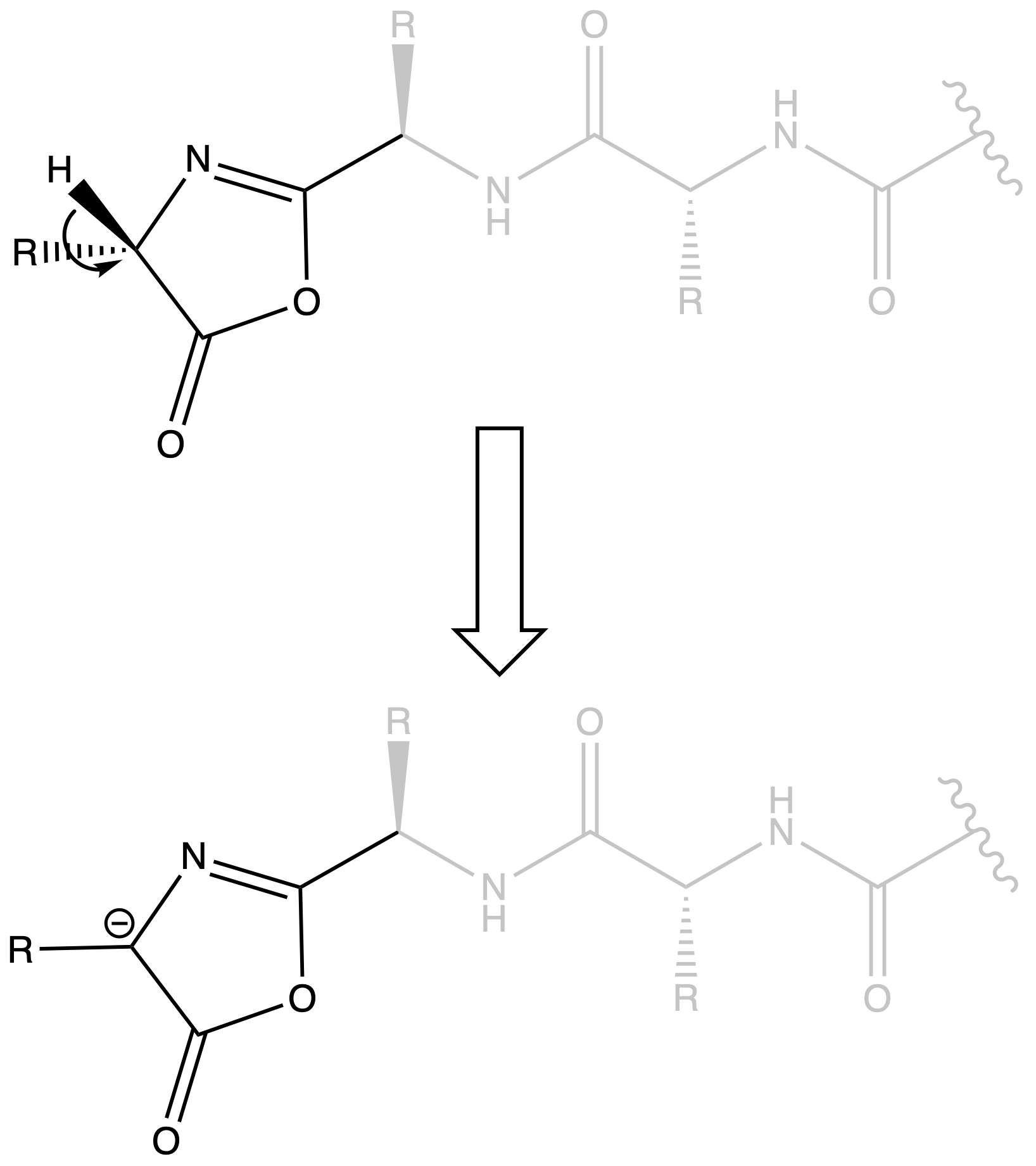
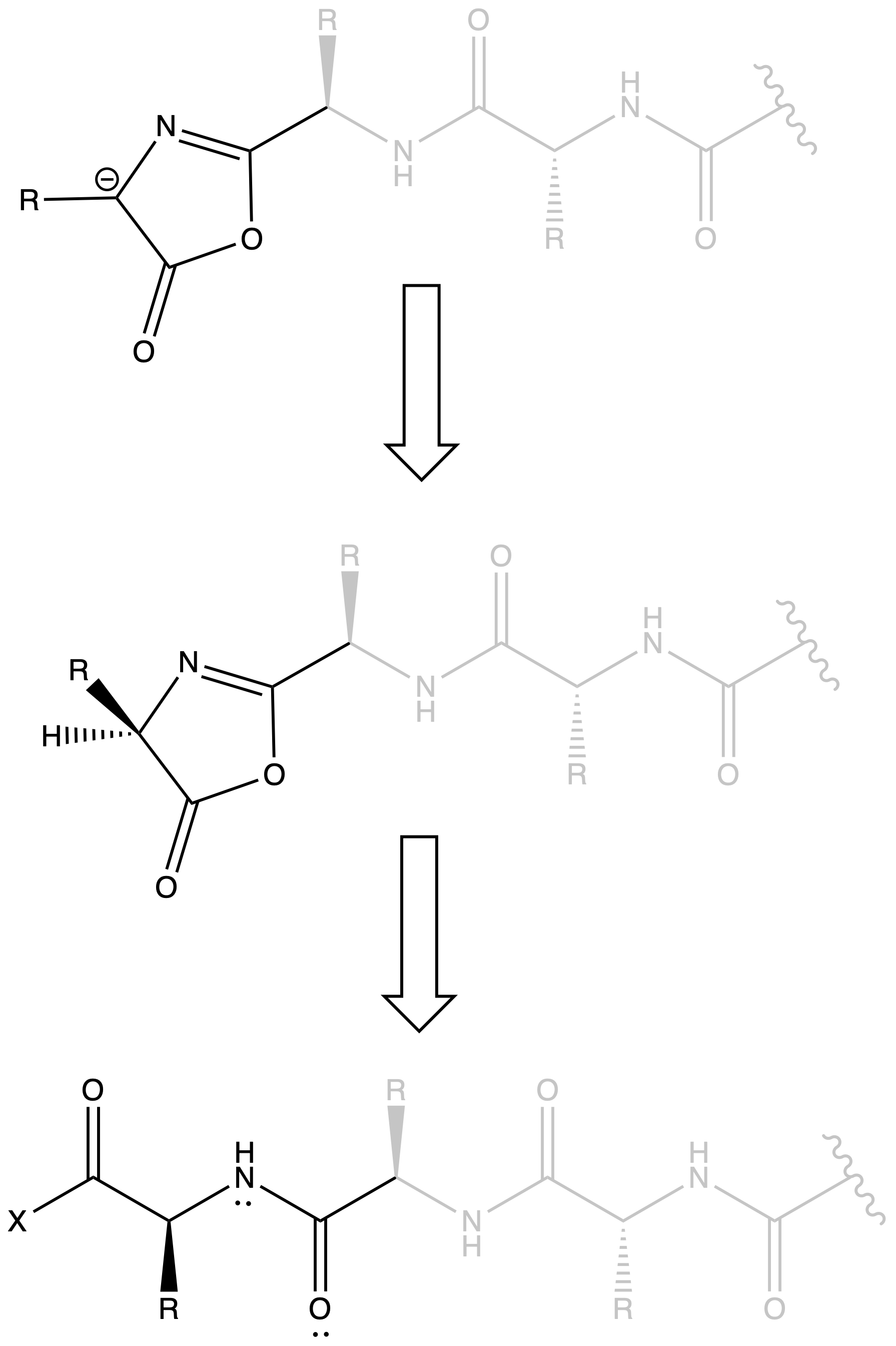
Oxazalone can result in Racemization
- When we have a peptide chain, oxalone can be formed near the terminal
- The lone pair of nitrogen is donated to the oxygen in an amide group, which means the oxygen is nucleophilic, allowing it to attack the terminal activated carboxyl group
- The hydrogen on oxazalone can be deprotonated as it will form an aromatic ring
- When the oxazalone gets protonated again, it may undergo racemization
The formation of oxazalone can be avoided if we can avoid having the amide group and the activated carboxyl group on the same molecule
- In N-to-C synthesis, both the activated carboxyl group and the amide group will be on the growing chain
- While in C-to-N synthesis, the activated carboxyl group will be on the free amino acid and the amide groups will be on the growing chain
We therefore choose the synthesize our peptides from the C-terminus to the N-terminus
Orthogonal Protecting Groups
For the backbone, we have to protect the C-terminus and the N-terminus
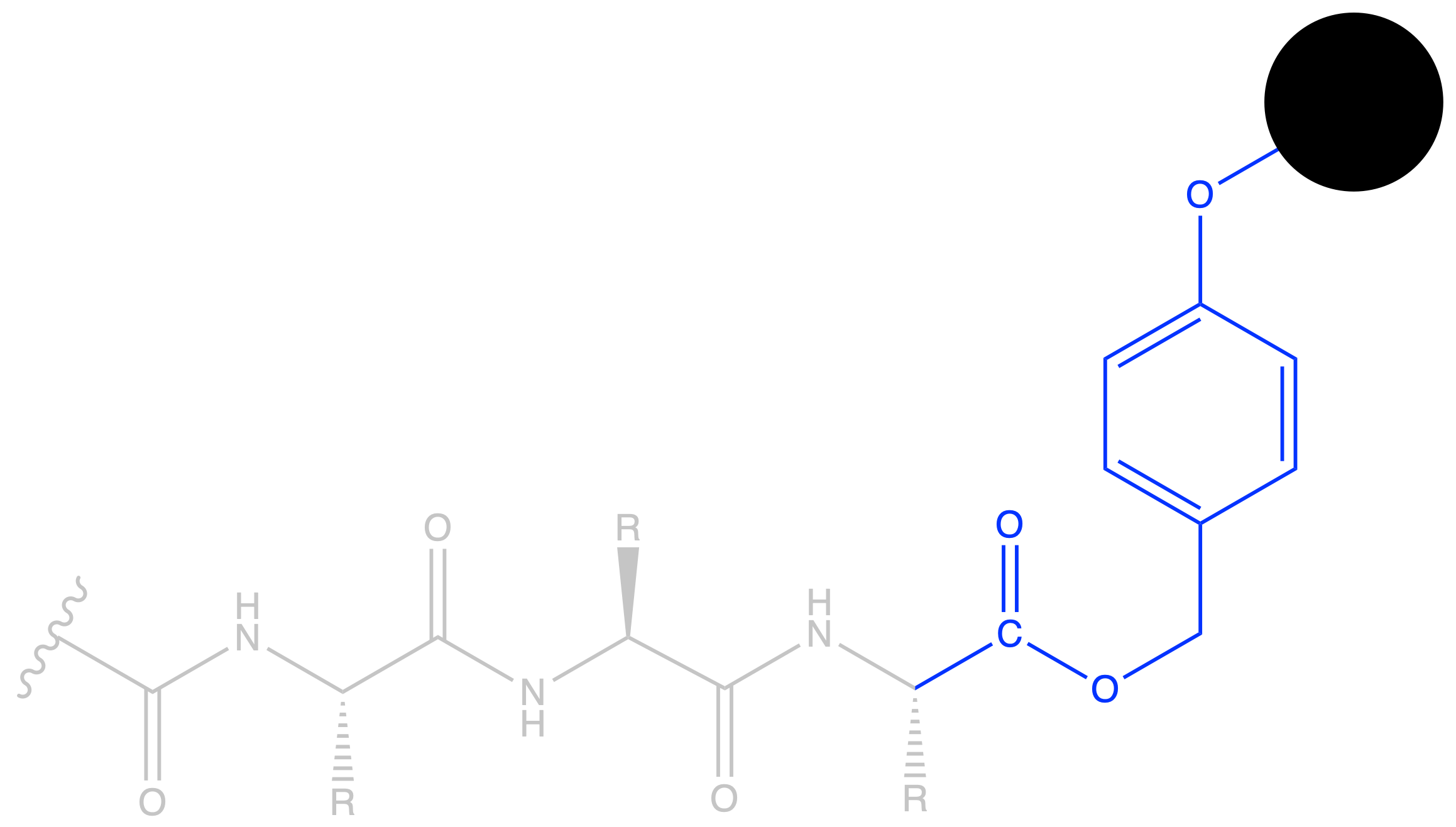
- Since we are synthesizing from the C-terminus, the C-terminus should already be linked to the resin. We choose PMB as the linker ( Wang resin ) so that it can be removed by acid at the end
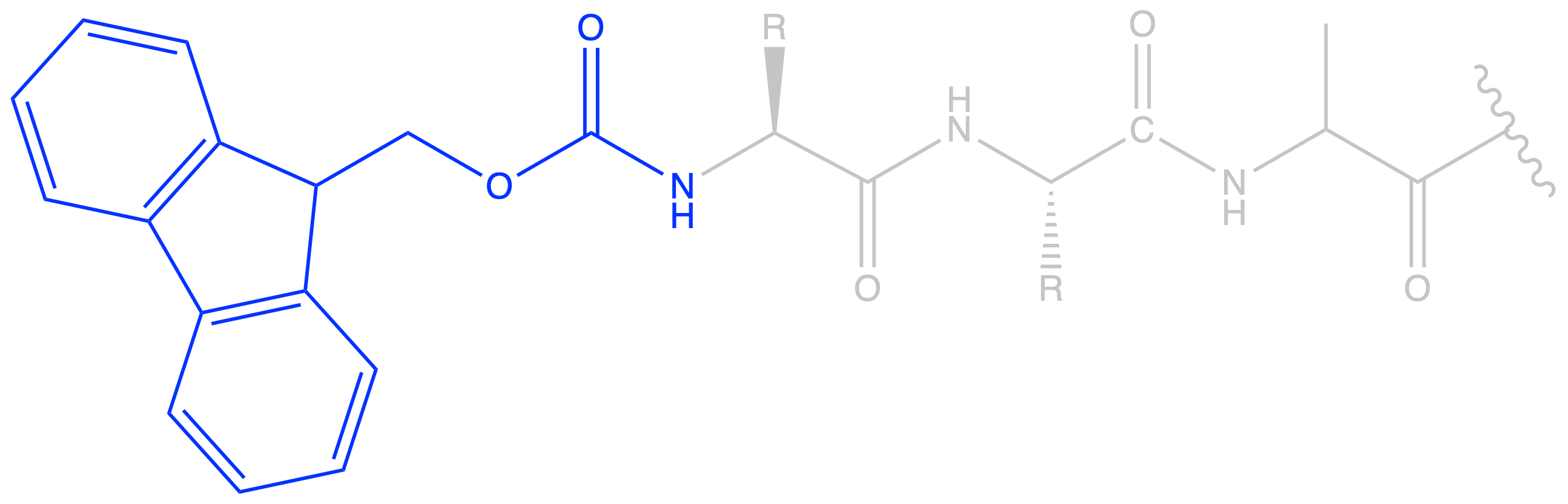
- We choose Fmoc as the protecting group for the amine as it is labile to base, making it orthogonal to PMB. Moreover, Fmoc is removed repeatedly with each round of coupling
Side chain functional groups also need protecting so they don’t interfere with coupling steps
- These should be ‘permanent’ protecting groups, meaning that they will removed only once at the end
- Notice the final step in the synthesis is removal of the peptide from the resin
- Ideally you want to minimise reaction steps, so this same step should also remove all side chain protecting groups
- Therefore you will want acid labile side chain protecting groups
Amide Bonds Formation
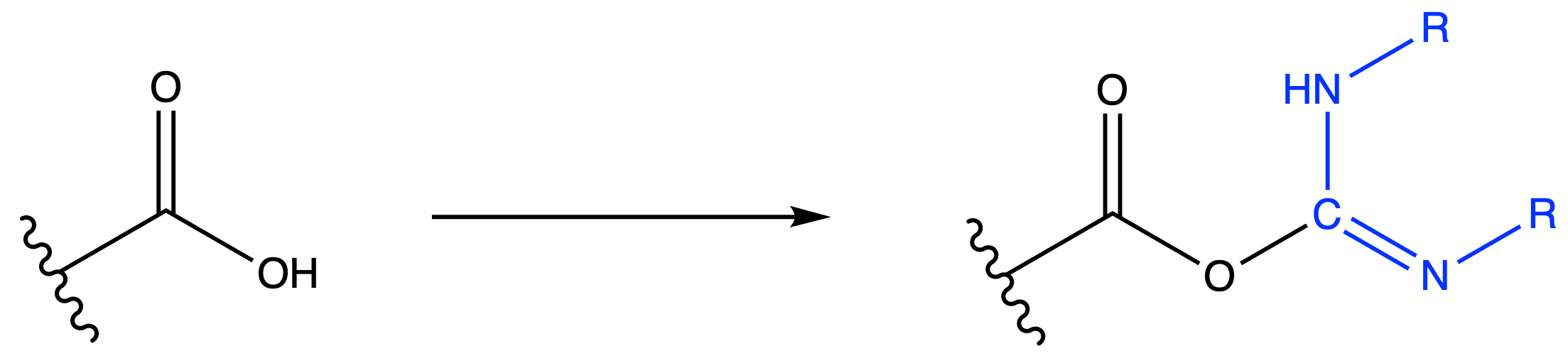
We need to activate the carboxyl group if we want amine to attack it
- You might first think of using an acid chloride, but this can be challenging as conditions to form this tend to be quite harsh and often incompatible with amino acid protecting groups
- Acid chlorides can also be too reactive and lead to side products
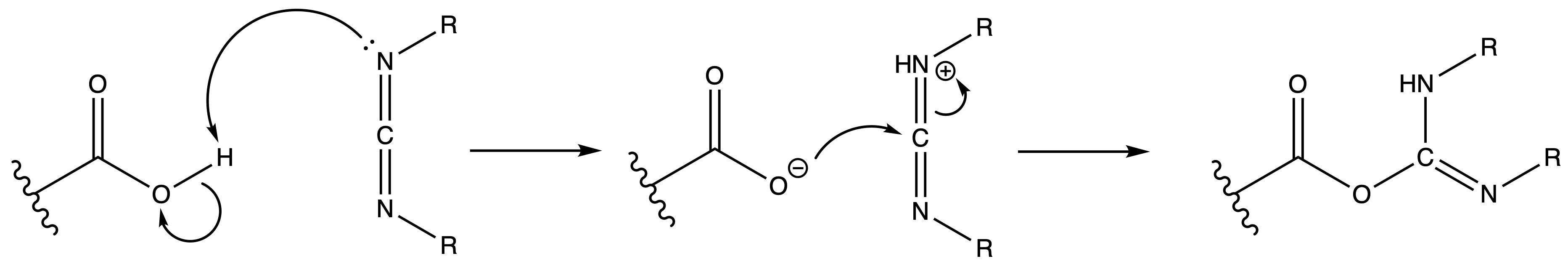
We instead use Carbodiimide coupling reagents to activate it
- Mechanism:
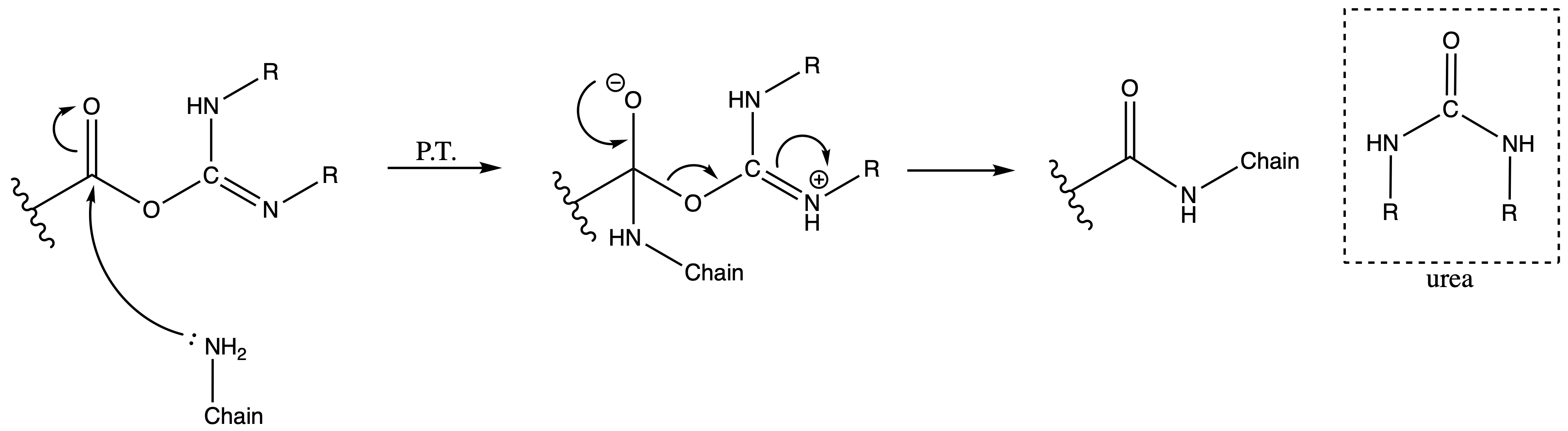
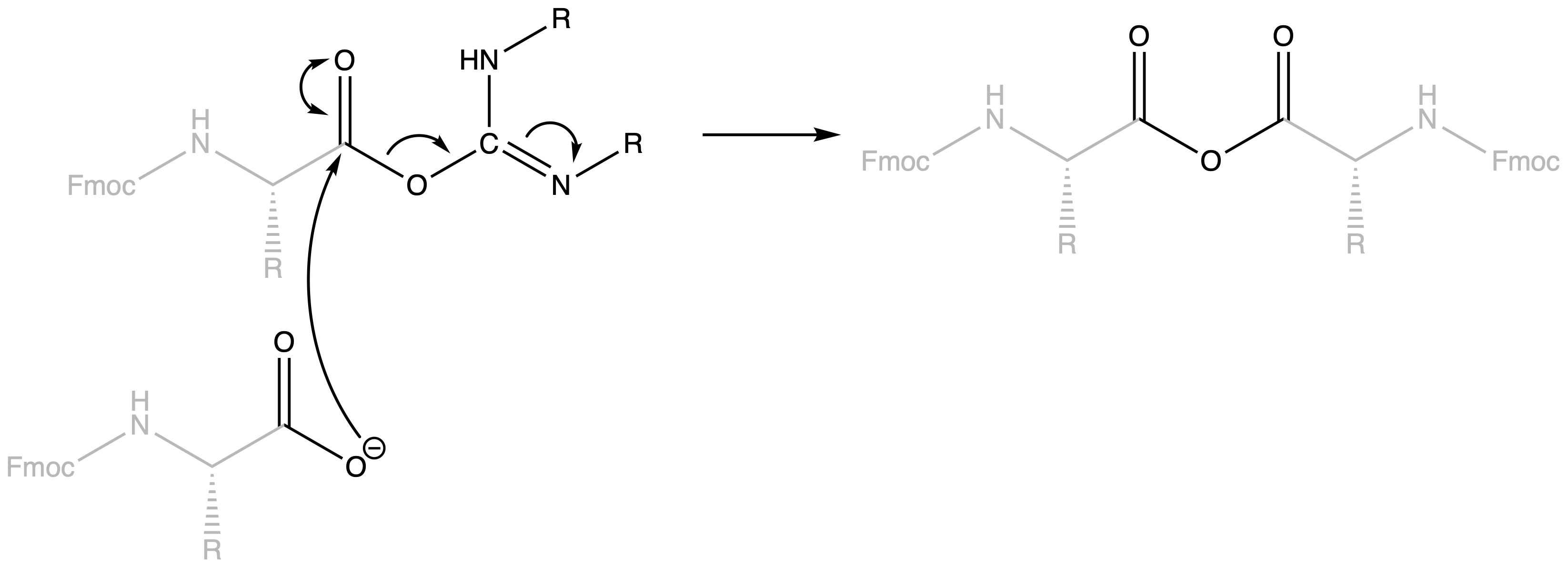
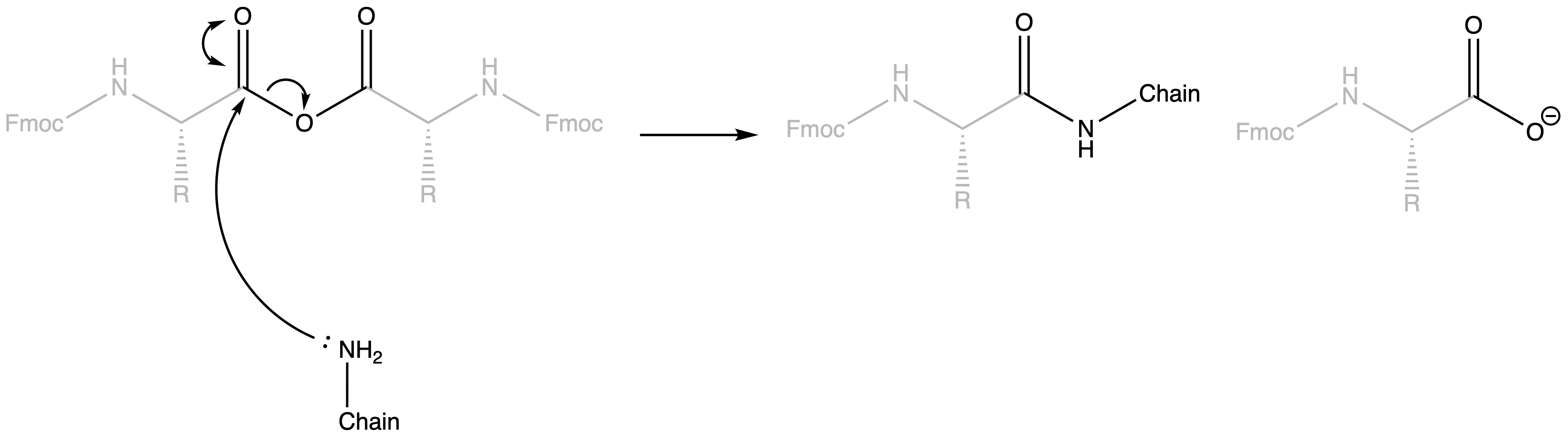
The activated carboxyl group can then form amide bonds
- Mechanism :
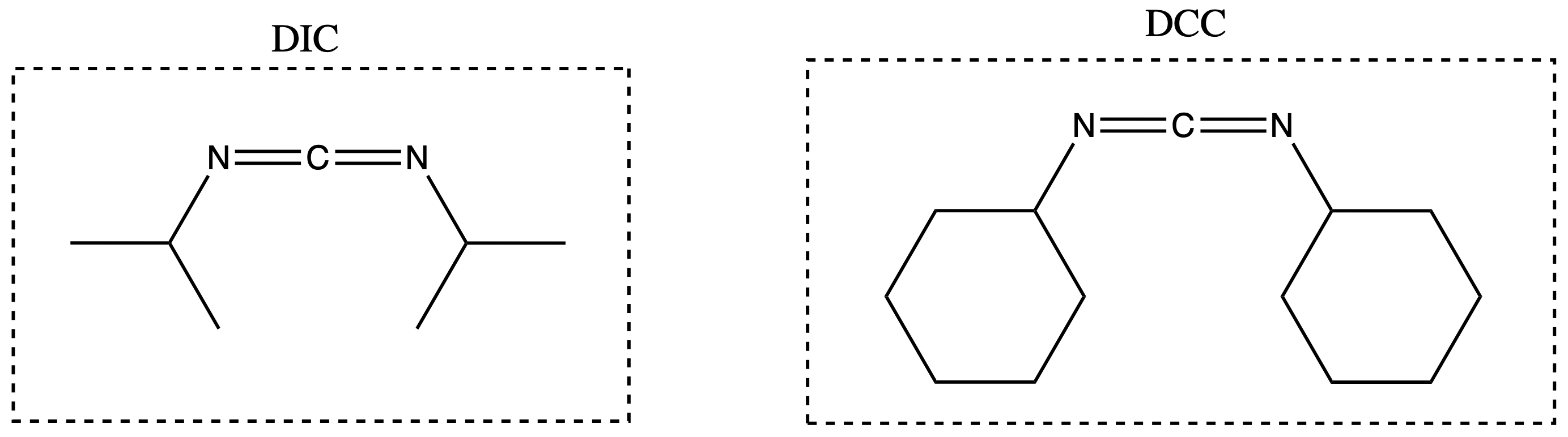
The group on the Carbodiimides is also important
- If = cyclohexyl, the urea precipitates. If = isopropyl, the urea stays soluble
- Insoluble biproduct helps drive reaction which is good for solutions phase reaction, but precipitate is bad for solid phase reaction so DIC is usually used
Unwanted side reactions can happen when we activate the group with carbodiimides
- Anhydride can be formed in the process
- Anhydride is quite reactive, so amide will still be formed subsequently
- However, this little maneuver will cost us 2 eq. acid per amide bond
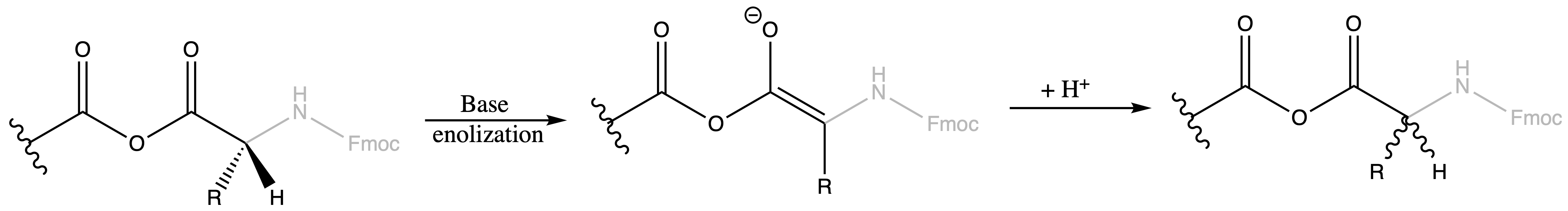
- Moreover, we can also get racemisation by removal of the alpha-proton
Anhydride formation and amino acid racemisation can be reduced by tuning the reactivity of the isoacyl urea intermediate
¶ Carbohydrates
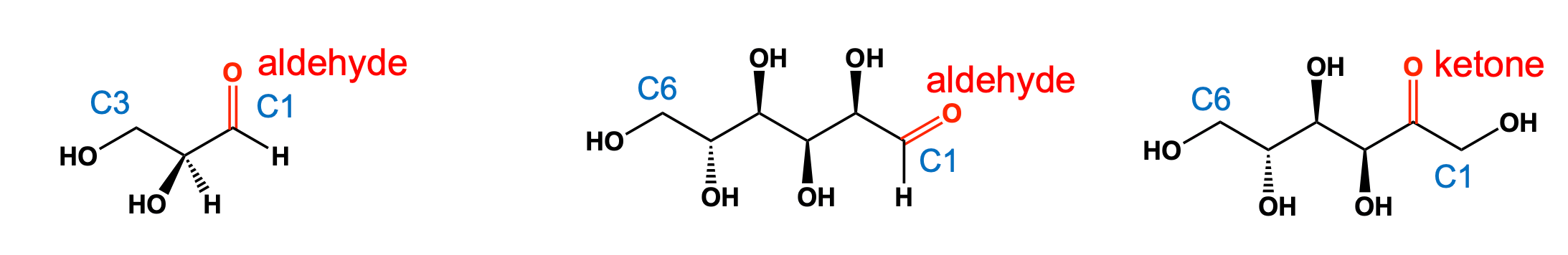
Carbohydrates are polyhydroxy aldehydes or ketones or compounds that yield such entities upon hydrolysis
- Numbering: C1 closest to aldehyde/ketone
¶ Cyclic Carbohydrates
Carbohydrates are often cyclic
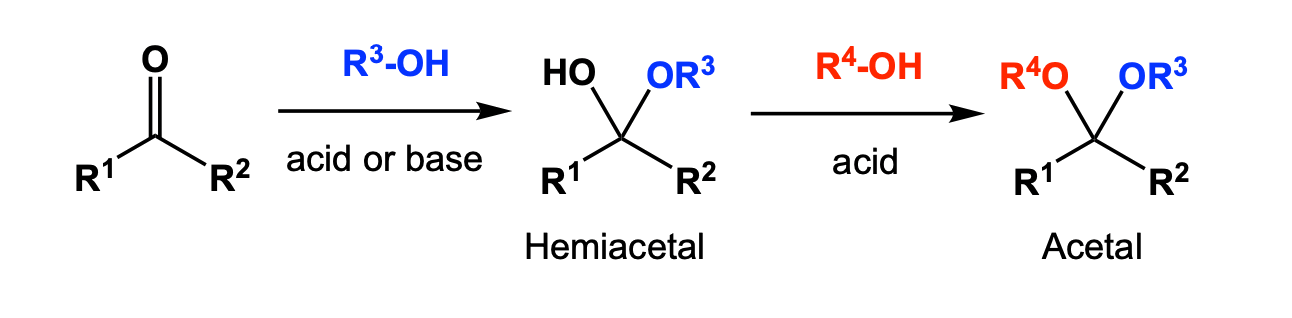
- Carbonyls can be converted to Hemiacetal and Acetal
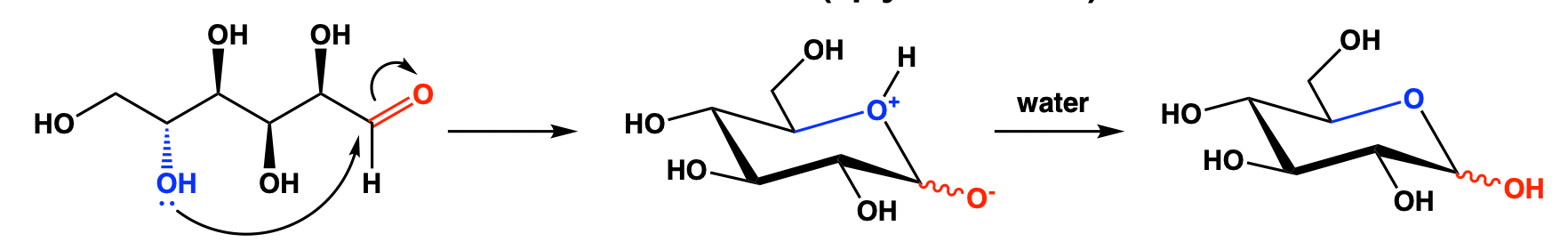
Mechanism:
Carbohydrates often contain both carbonyl and hydroxyl groups, so they can undergo self-cyclisation